Pre Gene Modal
BGIBMGA007711
Annotation
PREDICTED:_tRNA_(adenine(58)-N(1))-methyltransferase_non-catalytic_subunit_TRM6_[Bombyx_mori]
Full name
tRNA (adenine(58)-N(1))-methyltransferase non-catalytic subunit TRM6
Location in the cell
Nuclear Reliability : 3.911
Sequence
CDS
ATGATAAGTAAAAGTAACAAAAAAGCCAGGGAATACACAGTGGAGCTAACTGACGAGGTGGTCAATCTAAAAGATGAGATTGAGATAAAATTATCTGGTAGTGATAATAGAAACATCGTTGATGATGGACTCTCACAGAAACTGACAGCTGCAGAAATCGAAGATCTGAAGAATGACGCTAATAGGGCTTCAGATATTATAGAATCTTTAATTACTAATTCAAATACCTTTCACAATAAAACAGAGTTTTCTCAGGAGAAGTATTTAAAGAAGAAAGAAAAGAAATATTTTGAATATTTACAAATATTAAAACCTAACCTAAGAACGATAGCAGAAATCATGTATAAATTAGAACCTGGTAAAGTTCAAAATTTAAGAATAGACACTTTGTCACAGATTATTACAACTGTGAATATTTATTGTGAGGGAAATCATTTATTATATGACTCTGGATCTAATGGTTTAGTTGCTGCAGCTTTATTAAGTTCTATTGGTCAAAATACTAATGGTAGACTAGTCCACATGCACCCCGGCAACATGTCTCAGAAACAAGCTCTCTTAGCTATGAATTTTCCAGAAGAACAACTTAAGAGATGTATTTCTGTTAATGTTTATTCTGCATTAAGACAATTTTATCAAGGTTGTGATACAAATTCCAATGAAAATATAAAAGAAATTCCTATCACAGAGTCAAATCAATTAAAAAGAAAATTAGTAGAAAATGACAATGAACCTAGAAGAAAAATTGCTAAACCTGATGAAGAAGTATCTGAACCAGTAATTGATGAATTAAAGGAAAATAGTAACAAAGGTGATCCAAAGATACCAAAATGGCATTTCGATAACATTTATGCTGCACAAATTCTTGAAAAAAAATTGGATTCATTGGTTATAACTTGTAAGGAAGATCCTCAAAATATATTTCTTGAATTAGTTAATTTTATCAAACCTGGCCGACCATTTGTTATTTACTATAATGTATCAGAGCCATTACAAAATCTATACTTAGCTTTAAAATCACAAAGTAATGTTGCAGCATTAAAACTCACAAGCAACTGGATGAGGAATTACCAGGTTTTGCCAGAACGAACACGTCCAGATGTTATTATGAATGGTGCTAGTGGGTATTTGTTATCAGGATATGTACTTAAATGA
Protein
MISKSNKKAREYTVELTDEVVNLKDEIEIKLSGSDNRNIVDDGLSQKLTAAEIEDLKNDANRASDIIESLITNSNTFHNKTEFSQEKYLKKKEKKYFEYLQILKPNLRTIAEIMYKLEPGKVQNLRIDTLSQIITTVNIYCEGNHLLYDSGSNGLVAAALLSSIGQNTNGRLVHMHPGNMSQKQALLAMNFPEEQLKRCISVNVYSALRQFYQGCDTNSNENIKEIPITESNQLKRKLVENDNEPRRKIAKPDEEVSEPVIDELKENSNKGDPKIPKWHFDNIYAAQILEKKLDSLVITCKEDPQNIFLELVNFIKPGRPFVIYYNVSEPLQNLYLALKSQSNVAALKLTSNWMRNYQVLPERTRPDVIMNGASGYLLSGYVLK
Summary
Description
Substrate-binding subunit of tRNA (adenine-N1-)-methyltransferase, which catalyzes the formation of N1-methyladenine at position 58 (m1A58) in initiator methionyl-tRNA.
Subunit
Heterotetramer.
Similarity
Belongs to the TRM6/GCD10 family.
Uniprot
A0A2A4K145
A0A3S2LII6
A0A0N1IQ72
S4NT16
A0A212F3X7
A0A194PJP7
+ More
A0A0L7LR26 U5ETR0 A0A1I8PC21 A0A1S4JVA7 A0A0L0C434 A0A1Q3EYB9 A0A1I8MWV6 A0A1I8MWV5 A0A1W4UC63 B4P0B8 A0A182M8S7 Q8MS69 B3MNZ2 B4KHU0 B4MUJ5 B4Q4Y4 A0A182W909 A0A182PTD4 A0A034WV42 A0A3B0JIU2 A0A182YMS9 B4HXU1 B0WVR7 A0A0R3NXC8 A0A182RR78 T1DL04 A0A2M4BMB7 B3N5T4 B5DV67 A0A2M4BN74 Q29PC8 B4GKI7 A0A182JTT5 A0A0M4E8P2 W8BEY1 Q17EU7 A0A0K8V879 B4JR99 W5JT56 A0A182KTK2 Q7PMA4 A0A2M4AR62 A0A182XJK6 A0A0A1WPD9 A0A1S4H2E6 A0A1A9W3X5 A0A182HRI4 A0A1A9V5P1 A0A182SAK3 B4LVE5 A0A182F3V8 A0A182UG57 A0A182IMM4 A0A182NAS2 A0A084W8N0 A0A182UUY2 A0A182GSG7 N6TZB4 U4US95 A0A1J1HHH1 D2A3U3 A0A336K501 A0A182Q2J5 A0A1B0DAR7 A0A0A1XTK9 E2BEK2 A0A087ZNU0 A0A2A3EPY7 A0A026WU60 A0A3L8DGQ8 E1ZYD0 A0A1B0G9W0 A0A0J7K997 T1GR17 K7ILS2 A0A1L8DG64 A0A195FUB5 A0A232FJ07 A0A0M8ZZM6 A0A0J7K9R4 A0A1B6HIR9 A0A1B6GEY5 E0VPC2 A0A1B6D8D8 A0A0J7K9T6 A0A0L7RB24 A0A069DUB4 A0A1B6KPM0 A0A310SMI0 A0A224XQ46
A0A0L7LR26 U5ETR0 A0A1I8PC21 A0A1S4JVA7 A0A0L0C434 A0A1Q3EYB9 A0A1I8MWV6 A0A1I8MWV5 A0A1W4UC63 B4P0B8 A0A182M8S7 Q8MS69 B3MNZ2 B4KHU0 B4MUJ5 B4Q4Y4 A0A182W909 A0A182PTD4 A0A034WV42 A0A3B0JIU2 A0A182YMS9 B4HXU1 B0WVR7 A0A0R3NXC8 A0A182RR78 T1DL04 A0A2M4BMB7 B3N5T4 B5DV67 A0A2M4BN74 Q29PC8 B4GKI7 A0A182JTT5 A0A0M4E8P2 W8BEY1 Q17EU7 A0A0K8V879 B4JR99 W5JT56 A0A182KTK2 Q7PMA4 A0A2M4AR62 A0A182XJK6 A0A0A1WPD9 A0A1S4H2E6 A0A1A9W3X5 A0A182HRI4 A0A1A9V5P1 A0A182SAK3 B4LVE5 A0A182F3V8 A0A182UG57 A0A182IMM4 A0A182NAS2 A0A084W8N0 A0A182UUY2 A0A182GSG7 N6TZB4 U4US95 A0A1J1HHH1 D2A3U3 A0A336K501 A0A182Q2J5 A0A1B0DAR7 A0A0A1XTK9 E2BEK2 A0A087ZNU0 A0A2A3EPY7 A0A026WU60 A0A3L8DGQ8 E1ZYD0 A0A1B0G9W0 A0A0J7K997 T1GR17 K7ILS2 A0A1L8DG64 A0A195FUB5 A0A232FJ07 A0A0M8ZZM6 A0A0J7K9R4 A0A1B6HIR9 A0A1B6GEY5 E0VPC2 A0A1B6D8D8 A0A0J7K9T6 A0A0L7RB24 A0A069DUB4 A0A1B6KPM0 A0A310SMI0 A0A224XQ46
Pubmed
26354079
23622113
22118469
26227816
26108605
25315136
+ More
17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25348373 25244985 15632085 24495485 17510324 20920257 23761445 20966253 12364791 25830018 24438588 26483478 23537049 18362917 19820115 20798317 24508170 30249741 20075255 28648823 20566863 26334808
17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25348373 25244985 15632085 24495485 17510324 20920257 23761445 20966253 12364791 25830018 24438588 26483478 23537049 18362917 19820115 20798317 24508170 30249741 20075255 28648823 20566863 26334808
EMBL
NWSH01000302
PCG77614.1
RSAL01000103
RVE47404.1
KQ459937
KPJ19210.1
+ More
GAIX01013827 JAA78733.1 AGBW02010474 OWR48448.1 KQ459602 KPI93253.1 JTDY01000275 KOB77953.1 GANO01002673 JAB57198.1 JRES01000933 KNC27118.1 GFDL01014755 JAV20290.1 CM000157 EDW87876.1 AXCM01000771 AE014134 AY119050 AAF52404.2 AAM50910.1 CH902620 EDV32179.1 CH933807 EDW13377.1 CH963857 EDW76190.1 CM000361 CM002910 EDX03976.1 KMY88547.1 GAKP01000877 JAC58075.1 OUUW01000006 SPP82344.1 CH480818 EDW51871.1 DS232131 EDS35674.1 CH675624 KRT05736.1 GAMD01000681 JAB00910.1 GGFJ01005086 MBW54227.1 CH954177 EDV59093.1 EDY71805.1 GGFJ01005087 MBW54228.1 CH379058 EAL34365.2 CH479184 EDW37153.1 CP012523 ALC38414.1 GAMC01018266 JAB88289.1 CH477279 EAT45004.1 GDHF01017248 JAI35066.1 CH916372 EDV99429.1 ADMH02000223 ETN67326.1 AAAB01008980 EAA14079.5 GGFK01009943 MBW43264.1 GBXI01013575 GBXI01000449 JAD00717.1 JAD13843.1 APCN01001706 CH940649 EDW63324.1 ATLV01021496 KE525319 KFB46574.1 JXUM01084972 KQ563470 KXJ73785.1 APGK01055165 KB741261 ENN71577.1 KB632344 ERL93046.1 CVRI01000004 CRK87453.1 KQ971348 EFA04885.1 UFQS01000101 UFQT01000101 SSW99587.1 SSX19967.1 AXCN02000660 AJVK01013383 GBXI01000064 JAD14228.1 GL447817 EFN85892.1 KZ288204 PBC33296.1 KK107109 EZA59176.1 QOIP01000008 RLU19614.1 GL435204 EFN73829.1 CCAG010015991 LBMM01011190 KMQ86983.1 CAQQ02197427 GFDF01008641 JAV05443.1 KQ981264 KYN44048.1 NNAY01000126 OXU30726.1 KQ435798 KOX73346.1 LBMM01011196 KMQ86976.1 GECU01033251 JAS74455.1 GECZ01008780 JAS60989.1 DS235363 EEB15228.1 GEDC01015340 JAS21958.1 LBMM01011146 KMQ87001.1 KQ414618 KOC67941.1 GBGD01001538 JAC87351.1 GEBQ01026803 JAT13174.1 KQ762329 OAD55946.1 GFTR01006163 JAW10263.1
GAIX01013827 JAA78733.1 AGBW02010474 OWR48448.1 KQ459602 KPI93253.1 JTDY01000275 KOB77953.1 GANO01002673 JAB57198.1 JRES01000933 KNC27118.1 GFDL01014755 JAV20290.1 CM000157 EDW87876.1 AXCM01000771 AE014134 AY119050 AAF52404.2 AAM50910.1 CH902620 EDV32179.1 CH933807 EDW13377.1 CH963857 EDW76190.1 CM000361 CM002910 EDX03976.1 KMY88547.1 GAKP01000877 JAC58075.1 OUUW01000006 SPP82344.1 CH480818 EDW51871.1 DS232131 EDS35674.1 CH675624 KRT05736.1 GAMD01000681 JAB00910.1 GGFJ01005086 MBW54227.1 CH954177 EDV59093.1 EDY71805.1 GGFJ01005087 MBW54228.1 CH379058 EAL34365.2 CH479184 EDW37153.1 CP012523 ALC38414.1 GAMC01018266 JAB88289.1 CH477279 EAT45004.1 GDHF01017248 JAI35066.1 CH916372 EDV99429.1 ADMH02000223 ETN67326.1 AAAB01008980 EAA14079.5 GGFK01009943 MBW43264.1 GBXI01013575 GBXI01000449 JAD00717.1 JAD13843.1 APCN01001706 CH940649 EDW63324.1 ATLV01021496 KE525319 KFB46574.1 JXUM01084972 KQ563470 KXJ73785.1 APGK01055165 KB741261 ENN71577.1 KB632344 ERL93046.1 CVRI01000004 CRK87453.1 KQ971348 EFA04885.1 UFQS01000101 UFQT01000101 SSW99587.1 SSX19967.1 AXCN02000660 AJVK01013383 GBXI01000064 JAD14228.1 GL447817 EFN85892.1 KZ288204 PBC33296.1 KK107109 EZA59176.1 QOIP01000008 RLU19614.1 GL435204 EFN73829.1 CCAG010015991 LBMM01011190 KMQ86983.1 CAQQ02197427 GFDF01008641 JAV05443.1 KQ981264 KYN44048.1 NNAY01000126 OXU30726.1 KQ435798 KOX73346.1 LBMM01011196 KMQ86976.1 GECU01033251 JAS74455.1 GECZ01008780 JAS60989.1 DS235363 EEB15228.1 GEDC01015340 JAS21958.1 LBMM01011146 KMQ87001.1 KQ414618 KOC67941.1 GBGD01001538 JAC87351.1 GEBQ01026803 JAT13174.1 KQ762329 OAD55946.1 GFTR01006163 JAW10263.1
Proteomes
UP000218220
UP000283053
UP000053240
UP000007151
UP000053268
UP000037510
+ More
UP000095300 UP000037069 UP000095301 UP000192221 UP000002282 UP000075883 UP000000803 UP000007801 UP000009192 UP000007798 UP000000304 UP000075920 UP000075885 UP000268350 UP000076408 UP000001292 UP000002320 UP000001819 UP000075900 UP000008711 UP000008744 UP000075881 UP000092553 UP000008820 UP000001070 UP000000673 UP000075882 UP000007062 UP000076407 UP000091820 UP000075840 UP000078200 UP000075901 UP000008792 UP000069272 UP000075902 UP000075880 UP000075884 UP000030765 UP000075903 UP000069940 UP000249989 UP000019118 UP000030742 UP000183832 UP000007266 UP000075886 UP000092462 UP000008237 UP000005203 UP000242457 UP000053097 UP000279307 UP000000311 UP000092444 UP000036403 UP000015102 UP000002358 UP000078541 UP000215335 UP000053105 UP000009046 UP000053825
UP000095300 UP000037069 UP000095301 UP000192221 UP000002282 UP000075883 UP000000803 UP000007801 UP000009192 UP000007798 UP000000304 UP000075920 UP000075885 UP000268350 UP000076408 UP000001292 UP000002320 UP000001819 UP000075900 UP000008711 UP000008744 UP000075881 UP000092553 UP000008820 UP000001070 UP000000673 UP000075882 UP000007062 UP000076407 UP000091820 UP000075840 UP000078200 UP000075901 UP000008792 UP000069272 UP000075902 UP000075880 UP000075884 UP000030765 UP000075903 UP000069940 UP000249989 UP000019118 UP000030742 UP000183832 UP000007266 UP000075886 UP000092462 UP000008237 UP000005203 UP000242457 UP000053097 UP000279307 UP000000311 UP000092444 UP000036403 UP000015102 UP000002358 UP000078541 UP000215335 UP000053105 UP000009046 UP000053825
PRIDE
Pfam
Interpro
IPR017423
TRM6
+ More
IPR029021 Prot-tyrosine_phosphatase-like
IPR016130 Tyr_Pase_AS
IPR020095 PsdUridine_synth_TruA_C
IPR000387 TYR_PHOSPHATASE_dom
IPR020103 PsdUridine_synth_cat_dom_sf
IPR020422 TYR_PHOSPHATASE_DUAL_dom
IPR000340 Dual-sp_phosphatase_cat-dom
IPR020097 PsdUridine_synth_TruA_a/b_dom
IPR036034 PDZ_sf
IPR029058 AB_hydrolase
IPR001478 PDZ
IPR013094 AB_hydrolase_3
IPR036972 Cyt_c_oxidase_su5b_sf
IPR005345 PHF5
IPR002124 Cyt_c_oxidase_su5b
IPR029021 Prot-tyrosine_phosphatase-like
IPR016130 Tyr_Pase_AS
IPR020095 PsdUridine_synth_TruA_C
IPR000387 TYR_PHOSPHATASE_dom
IPR020103 PsdUridine_synth_cat_dom_sf
IPR020422 TYR_PHOSPHATASE_DUAL_dom
IPR000340 Dual-sp_phosphatase_cat-dom
IPR020097 PsdUridine_synth_TruA_a/b_dom
IPR036034 PDZ_sf
IPR029058 AB_hydrolase
IPR001478 PDZ
IPR013094 AB_hydrolase_3
IPR036972 Cyt_c_oxidase_su5b_sf
IPR005345 PHF5
IPR002124 Cyt_c_oxidase_su5b
Gene 3D
ProteinModelPortal
A0A2A4K145
A0A3S2LII6
A0A0N1IQ72
S4NT16
A0A212F3X7
A0A194PJP7
+ More
A0A0L7LR26 U5ETR0 A0A1I8PC21 A0A1S4JVA7 A0A0L0C434 A0A1Q3EYB9 A0A1I8MWV6 A0A1I8MWV5 A0A1W4UC63 B4P0B8 A0A182M8S7 Q8MS69 B3MNZ2 B4KHU0 B4MUJ5 B4Q4Y4 A0A182W909 A0A182PTD4 A0A034WV42 A0A3B0JIU2 A0A182YMS9 B4HXU1 B0WVR7 A0A0R3NXC8 A0A182RR78 T1DL04 A0A2M4BMB7 B3N5T4 B5DV67 A0A2M4BN74 Q29PC8 B4GKI7 A0A182JTT5 A0A0M4E8P2 W8BEY1 Q17EU7 A0A0K8V879 B4JR99 W5JT56 A0A182KTK2 Q7PMA4 A0A2M4AR62 A0A182XJK6 A0A0A1WPD9 A0A1S4H2E6 A0A1A9W3X5 A0A182HRI4 A0A1A9V5P1 A0A182SAK3 B4LVE5 A0A182F3V8 A0A182UG57 A0A182IMM4 A0A182NAS2 A0A084W8N0 A0A182UUY2 A0A182GSG7 N6TZB4 U4US95 A0A1J1HHH1 D2A3U3 A0A336K501 A0A182Q2J5 A0A1B0DAR7 A0A0A1XTK9 E2BEK2 A0A087ZNU0 A0A2A3EPY7 A0A026WU60 A0A3L8DGQ8 E1ZYD0 A0A1B0G9W0 A0A0J7K997 T1GR17 K7ILS2 A0A1L8DG64 A0A195FUB5 A0A232FJ07 A0A0M8ZZM6 A0A0J7K9R4 A0A1B6HIR9 A0A1B6GEY5 E0VPC2 A0A1B6D8D8 A0A0J7K9T6 A0A0L7RB24 A0A069DUB4 A0A1B6KPM0 A0A310SMI0 A0A224XQ46
A0A0L7LR26 U5ETR0 A0A1I8PC21 A0A1S4JVA7 A0A0L0C434 A0A1Q3EYB9 A0A1I8MWV6 A0A1I8MWV5 A0A1W4UC63 B4P0B8 A0A182M8S7 Q8MS69 B3MNZ2 B4KHU0 B4MUJ5 B4Q4Y4 A0A182W909 A0A182PTD4 A0A034WV42 A0A3B0JIU2 A0A182YMS9 B4HXU1 B0WVR7 A0A0R3NXC8 A0A182RR78 T1DL04 A0A2M4BMB7 B3N5T4 B5DV67 A0A2M4BN74 Q29PC8 B4GKI7 A0A182JTT5 A0A0M4E8P2 W8BEY1 Q17EU7 A0A0K8V879 B4JR99 W5JT56 A0A182KTK2 Q7PMA4 A0A2M4AR62 A0A182XJK6 A0A0A1WPD9 A0A1S4H2E6 A0A1A9W3X5 A0A182HRI4 A0A1A9V5P1 A0A182SAK3 B4LVE5 A0A182F3V8 A0A182UG57 A0A182IMM4 A0A182NAS2 A0A084W8N0 A0A182UUY2 A0A182GSG7 N6TZB4 U4US95 A0A1J1HHH1 D2A3U3 A0A336K501 A0A182Q2J5 A0A1B0DAR7 A0A0A1XTK9 E2BEK2 A0A087ZNU0 A0A2A3EPY7 A0A026WU60 A0A3L8DGQ8 E1ZYD0 A0A1B0G9W0 A0A0J7K997 T1GR17 K7ILS2 A0A1L8DG64 A0A195FUB5 A0A232FJ07 A0A0M8ZZM6 A0A0J7K9R4 A0A1B6HIR9 A0A1B6GEY5 E0VPC2 A0A1B6D8D8 A0A0J7K9T6 A0A0L7RB24 A0A069DUB4 A0A1B6KPM0 A0A310SMI0 A0A224XQ46
PDB
5CD1
E-value=9.59712e-42,
Score=428
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus
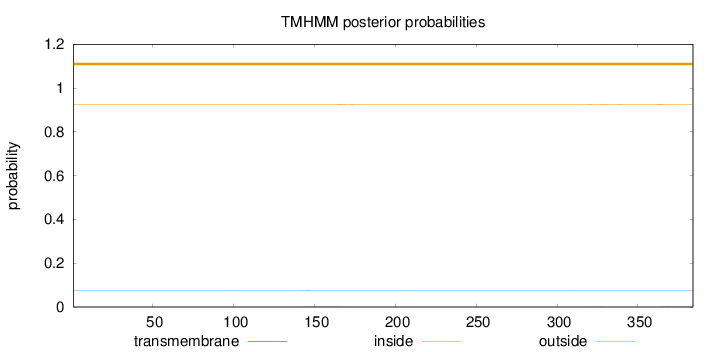
Length:
384
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.06427
Exp number, first 60 AAs:
0
Total prob of N-in:
0.07595
outside
1 - 384
Population Genetic Test Statistics
Pi
209.678222
Theta
186.149408
Tajima's D
0.216685
CLR
11.839034
CSRT
0.435728213589321
Interpretation
Uncertain
