Gene
KWMTBOMO08895
Annotation
PREDICTED:_anoctamin-5-like_[Papilio_machaon]
Full name
Anoctamin
Location in the cell
Cytoplasmic Reliability : 1.582 Nuclear Reliability : 1.379
Sequence
CDS
ATGGAAAACAAGCCCAAAGTTTTTACTGGATATTTTGCGGACGGCGTCAGGCGTATAGATTTAGTTTTGGTATTACGCGATGATGGAAATTCGCAAATTGAGGGCATCACTAATACTTTTCTCCTGAACGCTTTGAAGCTTGGTCTCGAAATAGAGATGGAGCATGGAACGATGAAACGACATAGAGACTTAATTTTTGCAAAAGTCCATGCACCGGACTCATTAATACGACAATACGCAGAAATTTTCAGTATTCAAAGACATTTCAAAGACAGTCATGTTGATTTCATAAACGCTGAGAATTCGTTCATGAAATATTTTAAAATGATAATGCCTTTCTTTTTTAACAGACGCGAAAGAGAATGGATCAAGTTGGTGCGGTAA
Protein
MENKPKVFTGYFADGVRRIDLVLVLRDDGNSQIEGITNTFLLNALKLGLEIEMEHGTMKRHRDLIFAKVHAPDSLIRQYAEIFSIQRHFKDSHVDFINAENSFMKYFKMIMPFFFNRREREWIKLVR
Summary
Similarity
Belongs to the anoctamin family.
Feature
chain Anoctamin
Uniprot
A0A0N1PHC9
A0A194PIV5
A0A212FIK9
A0A212F4Z3
A0A0N1IAC1
A0A0N0PE47
+ More
A0A0N1IJG4 A0A315V6U1 A0A0S7F3S0 A0A0S7F6X8 A0A0S7F0I6 A0A0S7F0J8 A0A0S7F2B9 A0A3B5QKS1 A0A0S7F0K1 A0A146SW80 A0A3B5M3J6 M3ZGE1 A0A0S7F8F2 A0A3B3X1S1 A0A0S7F5Z7 A0A0S7FAM4 A0A3B3X1P4 A0A146RFF4 A0A3B3X1N5 A0A087X9R6 A0A1A7XS53 A0A3B3VMQ6 A0A1A8V1B6 A0A3P9QHR5 A0A3Q2PWG4 A0A146NUQ8 A0A3Q3K045 A0A3P9QHS8 A0A3P9QHS3 A0A146NGE1 A0A1A8LBT2 A0A1A8NAJ3 A0A1A8A2U6 A0A1A8BTP4 A0A1A8CHP7 A0A3P9HUU8 A0A3P9HUS4 A0A3Q2ZK61 A0A3P9HV23 A0A3Q2ZY32 A0A3B4A541 A0A437DHS6 A0A3Q2ZHZ3 A0A3B3HFN8 A0A3B3HI13 A0A3P9H3W7 A0A1A8S082 A0A3B3CKH0 A0A3B3DT94 A0A3P9M2U9 A0A3P9H444 A0A3P9KLF0 H2LP37 A0A3P9KWV5 A0A3P9ITP7 A0A3B3IIG2 A0A3P9H3Z9 A0A3B3H499 A0A3P9M2S5 A0A3P9M364 A0A3B3HV27 A0A3P9M9K5 A0A3P9MA37 A0A3P9H3V3 A0A3B3IHD1 H2LMT2 A0A2I4D7B5 A0A2I4D7C2 A0A437D9P0 A0A3Q2E9F9 A0A3Q2D6H1 A0A2U9BC08 L5MGS7 A0A0P6EN60 A0A3Q3E4R9 I3J461 A0A3B4TYI0 A0A3B4YWQ9 I3J462 A0A3B4VAH1 A0A091JU70 A0A0F8ATW0
A0A0N1IJG4 A0A315V6U1 A0A0S7F3S0 A0A0S7F6X8 A0A0S7F0I6 A0A0S7F0J8 A0A0S7F2B9 A0A3B5QKS1 A0A0S7F0K1 A0A146SW80 A0A3B5M3J6 M3ZGE1 A0A0S7F8F2 A0A3B3X1S1 A0A0S7F5Z7 A0A0S7FAM4 A0A3B3X1P4 A0A146RFF4 A0A3B3X1N5 A0A087X9R6 A0A1A7XS53 A0A3B3VMQ6 A0A1A8V1B6 A0A3P9QHR5 A0A3Q2PWG4 A0A146NUQ8 A0A3Q3K045 A0A3P9QHS8 A0A3P9QHS3 A0A146NGE1 A0A1A8LBT2 A0A1A8NAJ3 A0A1A8A2U6 A0A1A8BTP4 A0A1A8CHP7 A0A3P9HUU8 A0A3P9HUS4 A0A3Q2ZK61 A0A3P9HV23 A0A3Q2ZY32 A0A3B4A541 A0A437DHS6 A0A3Q2ZHZ3 A0A3B3HFN8 A0A3B3HI13 A0A3P9H3W7 A0A1A8S082 A0A3B3CKH0 A0A3B3DT94 A0A3P9M2U9 A0A3P9H444 A0A3P9KLF0 H2LP37 A0A3P9KWV5 A0A3P9ITP7 A0A3B3IIG2 A0A3P9H3Z9 A0A3B3H499 A0A3P9M2S5 A0A3P9M364 A0A3B3HV27 A0A3P9M9K5 A0A3P9MA37 A0A3P9H3V3 A0A3B3IHD1 H2LMT2 A0A2I4D7B5 A0A2I4D7C2 A0A437D9P0 A0A3Q2E9F9 A0A3Q2D6H1 A0A2U9BC08 L5MGS7 A0A0P6EN60 A0A3Q3E4R9 I3J461 A0A3B4TYI0 A0A3B4YWQ9 I3J462 A0A3B4VAH1 A0A091JU70 A0A0F8ATW0
EMBL
KQ459937
KPJ19209.1
KQ459602
KPI93252.1
AGBW02008366
OWR53564.1
+ More
AGBW02010284 OWR48811.1 KPJ19207.1 KPJ19205.1 KPJ19211.1 NHOQ01002268 PWA19055.1 GBYX01470787 JAO10856.1 GBYX01470781 JAO10862.1 GBYX01470788 JAO10855.1 GBYX01470789 JAO10854.1 GBYX01470774 JAO10869.1 GBYX01470772 JAO10871.1 GCES01101565 JAQ84757.1 GBYX01470776 JAO10867.1 GBYX01470784 GBYX01470779 JAO10864.1 GBYX01470785 GBYX01470778 JAO10858.1 GCES01119642 JAQ66680.1 AYCK01005565 HADW01019274 SBP20674.1 HAEJ01012910 SBS53367.1 GCES01150838 JAQ35484.1 GCES01156216 JAQ30106.1 HAEF01003962 SBR41344.1 HAEH01000926 SBR65871.1 HADY01010010 SBP48495.1 HADZ01006633 SBP70574.1 HADZ01014632 HAEA01006237 SBP78573.1 CM012439 RVE74467.1 HAEH01020516 SBS10914.1 CM012442 RVE71756.1 CP026247 AWP01493.1 KB100321 ELK37571.1 GDIQ01073282 JAN21455.1 AERX01013796 KK533302 KFP28682.1 KQ042768 KKF10939.1
AGBW02010284 OWR48811.1 KPJ19207.1 KPJ19205.1 KPJ19211.1 NHOQ01002268 PWA19055.1 GBYX01470787 JAO10856.1 GBYX01470781 JAO10862.1 GBYX01470788 JAO10855.1 GBYX01470789 JAO10854.1 GBYX01470774 JAO10869.1 GBYX01470772 JAO10871.1 GCES01101565 JAQ84757.1 GBYX01470776 JAO10867.1 GBYX01470784 GBYX01470779 JAO10864.1 GBYX01470785 GBYX01470778 JAO10858.1 GCES01119642 JAQ66680.1 AYCK01005565 HADW01019274 SBP20674.1 HAEJ01012910 SBS53367.1 GCES01150838 JAQ35484.1 GCES01156216 JAQ30106.1 HAEF01003962 SBR41344.1 HAEH01000926 SBR65871.1 HADY01010010 SBP48495.1 HADZ01006633 SBP70574.1 HADZ01014632 HAEA01006237 SBP78573.1 CM012439 RVE74467.1 HAEH01020516 SBS10914.1 CM012442 RVE71756.1 CP026247 AWP01493.1 KB100321 ELK37571.1 GDIQ01073282 JAN21455.1 AERX01013796 KK533302 KFP28682.1 KQ042768 KKF10939.1
Proteomes
Interpro
IPR032394
Anoct_dimer
+ More
IPR036188 FAD/NAD-bd_sf
IPR007632 Anoctamin
IPR002018 CarbesteraseB
IPR029058 AB_hydrolase
IPR020846 MFS_dom
IPR011701 MFS
IPR036259 MFS_trans_sf
IPR012347 Ferritin-like
IPR031294 Anoctamin-5
IPR009078 Ferritin-like_SF
IPR016024 ARM-type_fold
IPR011989 ARM-like
IPR000225 Armadillo
IPR036188 FAD/NAD-bd_sf
IPR007632 Anoctamin
IPR002018 CarbesteraseB
IPR029058 AB_hydrolase
IPR020846 MFS_dom
IPR011701 MFS
IPR036259 MFS_trans_sf
IPR012347 Ferritin-like
IPR031294 Anoctamin-5
IPR009078 Ferritin-like_SF
IPR016024 ARM-type_fold
IPR011989 ARM-like
IPR000225 Armadillo
Gene 3D
CDD
ProteinModelPortal
A0A0N1PHC9
A0A194PIV5
A0A212FIK9
A0A212F4Z3
A0A0N1IAC1
A0A0N0PE47
+ More
A0A0N1IJG4 A0A315V6U1 A0A0S7F3S0 A0A0S7F6X8 A0A0S7F0I6 A0A0S7F0J8 A0A0S7F2B9 A0A3B5QKS1 A0A0S7F0K1 A0A146SW80 A0A3B5M3J6 M3ZGE1 A0A0S7F8F2 A0A3B3X1S1 A0A0S7F5Z7 A0A0S7FAM4 A0A3B3X1P4 A0A146RFF4 A0A3B3X1N5 A0A087X9R6 A0A1A7XS53 A0A3B3VMQ6 A0A1A8V1B6 A0A3P9QHR5 A0A3Q2PWG4 A0A146NUQ8 A0A3Q3K045 A0A3P9QHS8 A0A3P9QHS3 A0A146NGE1 A0A1A8LBT2 A0A1A8NAJ3 A0A1A8A2U6 A0A1A8BTP4 A0A1A8CHP7 A0A3P9HUU8 A0A3P9HUS4 A0A3Q2ZK61 A0A3P9HV23 A0A3Q2ZY32 A0A3B4A541 A0A437DHS6 A0A3Q2ZHZ3 A0A3B3HFN8 A0A3B3HI13 A0A3P9H3W7 A0A1A8S082 A0A3B3CKH0 A0A3B3DT94 A0A3P9M2U9 A0A3P9H444 A0A3P9KLF0 H2LP37 A0A3P9KWV5 A0A3P9ITP7 A0A3B3IIG2 A0A3P9H3Z9 A0A3B3H499 A0A3P9M2S5 A0A3P9M364 A0A3B3HV27 A0A3P9M9K5 A0A3P9MA37 A0A3P9H3V3 A0A3B3IHD1 H2LMT2 A0A2I4D7B5 A0A2I4D7C2 A0A437D9P0 A0A3Q2E9F9 A0A3Q2D6H1 A0A2U9BC08 L5MGS7 A0A0P6EN60 A0A3Q3E4R9 I3J461 A0A3B4TYI0 A0A3B4YWQ9 I3J462 A0A3B4VAH1 A0A091JU70 A0A0F8ATW0
A0A0N1IJG4 A0A315V6U1 A0A0S7F3S0 A0A0S7F6X8 A0A0S7F0I6 A0A0S7F0J8 A0A0S7F2B9 A0A3B5QKS1 A0A0S7F0K1 A0A146SW80 A0A3B5M3J6 M3ZGE1 A0A0S7F8F2 A0A3B3X1S1 A0A0S7F5Z7 A0A0S7FAM4 A0A3B3X1P4 A0A146RFF4 A0A3B3X1N5 A0A087X9R6 A0A1A7XS53 A0A3B3VMQ6 A0A1A8V1B6 A0A3P9QHR5 A0A3Q2PWG4 A0A146NUQ8 A0A3Q3K045 A0A3P9QHS8 A0A3P9QHS3 A0A146NGE1 A0A1A8LBT2 A0A1A8NAJ3 A0A1A8A2U6 A0A1A8BTP4 A0A1A8CHP7 A0A3P9HUU8 A0A3P9HUS4 A0A3Q2ZK61 A0A3P9HV23 A0A3Q2ZY32 A0A3B4A541 A0A437DHS6 A0A3Q2ZHZ3 A0A3B3HFN8 A0A3B3HI13 A0A3P9H3W7 A0A1A8S082 A0A3B3CKH0 A0A3B3DT94 A0A3P9M2U9 A0A3P9H444 A0A3P9KLF0 H2LP37 A0A3P9KWV5 A0A3P9ITP7 A0A3B3IIG2 A0A3P9H3Z9 A0A3B3H499 A0A3P9M2S5 A0A3P9M364 A0A3B3HV27 A0A3P9M9K5 A0A3P9MA37 A0A3P9H3V3 A0A3B3IHD1 H2LMT2 A0A2I4D7B5 A0A2I4D7C2 A0A437D9P0 A0A3Q2E9F9 A0A3Q2D6H1 A0A2U9BC08 L5MGS7 A0A0P6EN60 A0A3Q3E4R9 I3J461 A0A3B4TYI0 A0A3B4YWQ9 I3J462 A0A3B4VAH1 A0A091JU70 A0A0F8ATW0
PDB
6QPI
E-value=0.000649291,
Score=95
Ontologies
GO
PANTHER
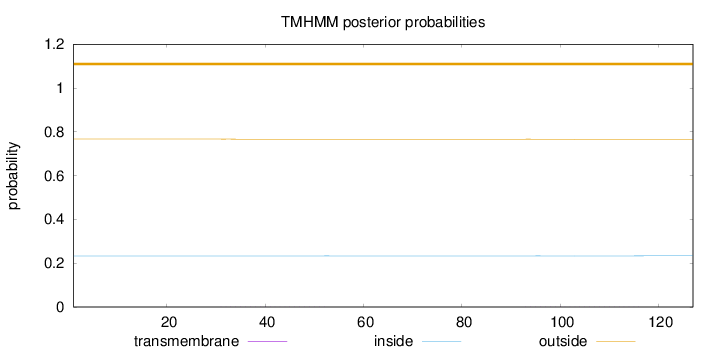
Topology
Subcellular location
Membrane
Length:
127
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03501
Exp number, first 60 AAs:
0.0101
Total prob of N-in:
0.23292
outside
1 - 127
Population Genetic Test Statistics
Pi
213.454889
Theta
152.276991
Tajima's D
1.220027
CLR
0
CSRT
0.721713914304285
Interpretation
Uncertain
