Pre Gene Modal
BGIBMGA007824
Annotation
PREDICTED:_succinyl-CoA_ligase_[GDP-forming]_subunit_beta?_mitochondrial-like_[Plutella_xylostella]
Full name
Succinate--CoA ligase [GDP-forming] subunit beta, mitochondrial
+ More
Succinate-CoA ligase subunit beta
Succinate-CoA ligase subunit beta
Alternative Name
GTP-specific succinyl-CoA synthetase subunit beta
Succinyl-CoA synthetase beta-G chain
Succinyl-CoA synthetase beta-G chain
Location in the cell
Cytoplasmic Reliability : 2.534
Sequence
CDS
ATGGCCGCTTTTAAGAATTTAAAAAGTTTTAAACTCATAAACCTTGCTTCGTCCACATATTGTCCACGAGTAATTTCAACAAGACACCTAAATCTTCAAGAGCATCATAGCAAAGATCTTCTGAGAAAATATCAGGTTTCCATTCAGGACTTCAGAATTATTGATTCCAAACTAGATACGAATGCCTTATCTGGTTTTAAAGCTGATGAATATGTAGTCAAGGCTCAGATTCTTGCTGGAGGACGAGGAAAAGGTCACTTTGACAATGGGTTTAAAGGGGGAGTTCATCTTACAAAAAACCGGGACAAGATTGTCGACTTAGCTAAAAATATGATAGGAAACAAGCTTATTACTAAACAAACTCCCAAAGAAGGTATTTTAGTAAATAAAGTTATGGTCGCAGAAAGTGTGAATATCAAAAGAGAGACATACTTTAGTATTGTAATGGAGAGAAGTTTTAATGGTGCAGCCATTGTAGCTTCTCCAGCTGGAGGTATGGATATAGAGGCAGTGGCTGAAAAAACCCCACATTTAGTGAAAACAGTTCCAGTAGATATTTTTGAAGGGATTAGTGACAAAGTTGCAAATGAAATAGCTGACTTTTTAGAATTCAAAGGACCACTAAAAAGCAAATGTGCTGAAGAAGTAAAAAAGCTATGGCAGTTGTTTTTGAAGGTGGATGCTACACAGCTTGAAATTAACCCTCTGGTTGAAACAGATGATGGACGTGTCATTTCAGTTGATGCCAAGATTAATTTTGATGACAATGCTAAGTTTAGACAAAAGGAGATCTTTGCACTTGATGATGTTTCCGAGACTGACCCACGTGAGAGAGAAGCAAATTCCCTCAATTTGGTGTACATTGAAATGGATGGAAGTATTGGCTGCATGGTTAATGGTGCAGGACTTGCCATGGCCACTATGGATATCATCACGCTGAATGGTGGCAAACCTGCTAACTTCCTAGACCTTGGAGGTGGAGTTGGTCAATCACAAGTTTCTGCAGCTTTGAAGATTTTAGAGTCTGATCCCAAAGTGAAGACGATTTTCGTCAATGTTTTTGGAGGTATTGTTAACTGTGCAACCATTGCAAATGGTATAGTTTCAGCATGCAGAGAGAATCCACCAAAACACCCCCTAGTCATTAGACTTGAAGGAACCAACTCTGGTGAAGCACGGAAAATCCTTGAGCAATCTGGTTTACCAGTTAAAATAATAAATGATGCCGACGAAGCTGCTAAAGCTGCTGTGAAGTTAGCACAGTGA
Protein
MAAFKNLKSFKLINLASSTYCPRVISTRHLNLQEHHSKDLLRKYQVSIQDFRIIDSKLDTNALSGFKADEYVVKAQILAGGRGKGHFDNGFKGGVHLTKNRDKIVDLAKNMIGNKLITKQTPKEGILVNKVMVAESVNIKRETYFSIVMERSFNGAAIVASPAGGMDIEAVAEKTPHLVKTVPVDIFEGISDKVANEIADFLEFKGPLKSKCAEEVKKLWQLFLKVDATQLEINPLVETDDGRVISVDAKINFDDNAKFRQKEIFALDDVSETDPREREANSLNLVYIEMDGSIGCMVNGAGLAMATMDIITLNGGKPANFLDLGGGVGQSQVSAALKILESDPKVKTIFVNVFGGIVNCATIANGIVSACRENPPKHPLVIRLEGTNSGEARKILEQSGLPVKIINDADEAAKAAVKLAQ
Summary
Description
GTP-specific succinyl-CoA synthetase functions in the citric acid cycle (TCA), coupling the hydrolysis of succinyl-CoA to the synthesis of GTP and thus represents the only step of substrate-level phosphorylation in the TCA. The beta subunit provides nucleotide specificity of the enzyme and binds the substrate succinate, while the binding sites for coenzyme A and phosphate are found in the alpha subunit.
Catalytic Activity
CoA + GTP + succinate = GDP + phosphate + succinyl-CoA
Cofactor
Mg(2+)
Subunit
Heterodimer of an alpha and a beta subunit. The beta subunit determines specificity for GTP.
Heterodimer of an alpha and a beta subunit.
Heterodimer of an alpha and a beta subunit.
Similarity
Belongs to the succinate/malate CoA ligase beta subunit family. GTP-specific subunit beta subfamily.
Belongs to the succinate/malate CoA ligase beta subunit family.
Belongs to the succinate/malate CoA ligase beta subunit family.
Feature
chain Succinate--CoA ligase [GDP-forming] subunit beta, mitochondrial
Uniprot
H9JE77
A0A2A4K021
A0A2H1VJW1
A0A194PKJ0
A0A0N0PEI7
A0A3S2P134
+ More
A0A0L7L8M9 A0A212FM66 N6U140 S4PRS6 A0A1Y1KDP3 A0A1W4XAU8 Q179P9 A0A182IRZ2 A0A182LNJ0 A0A182K4E2 A0A2S2NF83 A0A182UQ76 A0A182U3I9 A0A023EU71 A0A182XF24 A0A182HVJ2 D6W7F8 A0A2M4BP07 A0A0T6AXJ2 A0A2M4AKG0 A0A069DUK8 A0A2M4BNY9 A0A2H8TGC3 W5JGQ2 A0A084WQ81 A0A182H9I6 J9K8K5 A0A2M4BP30 A0A2M3ZHZ0 A0A182QGG2 Q7QA95 A0A182P3F3 A0A182FAR5 A0A182YJP7 R4G3Z4 B4LBD9 A0A0A9XS56 A0A1Q3FGM3 A0A0V0GC39 A0A224XCT7 A0A182W339 A0A182NCQ1 B0WEX9 A0A2P8YL05 B4PIU4 B4QRL3 A0A1W4VSI0 A0A0P5BPC8 B3NGC1 E9FY81 B4KVB1 A0A164QUM9 A0A0N8A5I7 B4HUG3 A0A0P5SR39 A0A0P5BL18 A0A023F898 Q9V470 A0A0P5E5E9 A0A0P5A382 A0A182LRP0 A0A0P6GNE8 A0A0N8ELI2 A0A2S2QVC8 B4MLR6 A0A0P5T9D6 A0A182R8H0 A0A232FCG8 U5EJ13 B3M3P7 K7IX86 A0A0A1X5E5 A0A026WJ19 A0A0L0C4E4 A0A0M5IYC5 B4IXS1 A0A3L8DXH5 A0A423U904 A0A3B0JAC1 A0A1I8MY09 A0A1B6H359 A0A0J7MY75 A0A2J7QJW3 A0A1B0GNB0 A0A2R7X8H7 A0A067RDN7 W8C579 A0A0K8TLA4 A0A1A9W8I5 A0A034VQW1 Q2LZM5 B4H3D0 E2A879 A0A1A9XRQ9
A0A0L7L8M9 A0A212FM66 N6U140 S4PRS6 A0A1Y1KDP3 A0A1W4XAU8 Q179P9 A0A182IRZ2 A0A182LNJ0 A0A182K4E2 A0A2S2NF83 A0A182UQ76 A0A182U3I9 A0A023EU71 A0A182XF24 A0A182HVJ2 D6W7F8 A0A2M4BP07 A0A0T6AXJ2 A0A2M4AKG0 A0A069DUK8 A0A2M4BNY9 A0A2H8TGC3 W5JGQ2 A0A084WQ81 A0A182H9I6 J9K8K5 A0A2M4BP30 A0A2M3ZHZ0 A0A182QGG2 Q7QA95 A0A182P3F3 A0A182FAR5 A0A182YJP7 R4G3Z4 B4LBD9 A0A0A9XS56 A0A1Q3FGM3 A0A0V0GC39 A0A224XCT7 A0A182W339 A0A182NCQ1 B0WEX9 A0A2P8YL05 B4PIU4 B4QRL3 A0A1W4VSI0 A0A0P5BPC8 B3NGC1 E9FY81 B4KVB1 A0A164QUM9 A0A0N8A5I7 B4HUG3 A0A0P5SR39 A0A0P5BL18 A0A023F898 Q9V470 A0A0P5E5E9 A0A0P5A382 A0A182LRP0 A0A0P6GNE8 A0A0N8ELI2 A0A2S2QVC8 B4MLR6 A0A0P5T9D6 A0A182R8H0 A0A232FCG8 U5EJ13 B3M3P7 K7IX86 A0A0A1X5E5 A0A026WJ19 A0A0L0C4E4 A0A0M5IYC5 B4IXS1 A0A3L8DXH5 A0A423U904 A0A3B0JAC1 A0A1I8MY09 A0A1B6H359 A0A0J7MY75 A0A2J7QJW3 A0A1B0GNB0 A0A2R7X8H7 A0A067RDN7 W8C579 A0A0K8TLA4 A0A1A9W8I5 A0A034VQW1 Q2LZM5 B4H3D0 E2A879 A0A1A9XRQ9
EC Number
6.2.1.4
6.2.1.-
6.2.1.-
Pubmed
19121390
26354079
26227816
22118469
23537049
23622113
+ More
28004739 17510324 20966253 24945155 18362917 19820115 26334808 20920257 23761445 24438588 26483478 12364791 14747013 17210077 25244985 17994087 25401762 26823975 29403074 17550304 22936249 21292972 25474469 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28648823 20075255 25830018 24508170 26108605 30249741 25315136 24845553 24495485 26369729 25348373 15632085 20798317
28004739 17510324 20966253 24945155 18362917 19820115 26334808 20920257 23761445 24438588 26483478 12364791 14747013 17210077 25244985 17994087 25401762 26823975 29403074 17550304 22936249 21292972 25474469 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28648823 20075255 25830018 24508170 26108605 30249741 25315136 24845553 24495485 26369729 25348373 15632085 20798317
EMBL
BABH01002718
NWSH01000302
PCG77611.1
ODYU01002957
SOQ41113.1
KQ459602
+ More
KPI93249.1 KQ459937 KPJ19203.1 RSAL01000005 RVE54341.1 JTDY01002339 KOB71636.1 AGBW02007668 OWR54779.1 APGK01047286 KB741077 KB632432 ENN74331.1 ERL95533.1 GAIX01000555 JAA92005.1 GEZM01087797 JAV58471.1 CH477347 EAT42975.1 EJY57594.1 GGMR01002817 MBY15436.1 GAPW01001639 JAC11959.1 APCN01005783 KQ971307 EFA11202.1 GGFJ01005666 MBW54807.1 LJIG01022565 KRT79897.1 GGFK01007955 MBW41276.1 GBGD01001537 JAC87352.1 GGFJ01005665 MBW54806.1 GFXV01001294 MBW13099.1 ADMH02001552 ETN62090.1 ATLV01025286 KE525379 KFB52375.1 JXUM01120885 KQ566440 KXJ70136.1 ABLF02025516 GGFJ01005664 MBW54805.1 GGFM01007382 MBW28133.1 AXCN02001180 AAAB01008898 EAA09142.4 ACPB03018838 GAHY01001790 JAA75720.1 CH940647 EDW69727.2 GBHO01021976 GBRD01005654 GBRD01005652 GBRD01005651 GDHC01000603 JAG21628.1 JAG60167.1 JAQ18026.1 GFDL01008351 JAV26694.1 GECL01001207 JAP04917.1 GFTR01006170 JAW10256.1 DS231912 EDS25837.1 PYGN01000519 PSN44935.1 CM000159 EDW93514.1 CM000363 CM002912 EDX09324.1 KMY97752.1 GDIP01182182 JAJ41220.1 CH954178 EDV51022.1 GL732527 EFX87492.1 CH933809 EDW18354.1 LRGB01002371 KZS08073.1 GDIP01180441 JAJ42961.1 CH480817 EDW50584.1 GDIP01136545 JAL67169.1 GDIP01183637 JAJ39765.1 GBBI01001166 JAC17546.1 AE014296 AY051980 AJ252069 AAF50771.1 AAK93404.1 CAB64384.1 GDIP01152595 JAJ70807.1 GDIP01218478 GDIP01216484 GDIP01204875 JAJ18527.1 AXCM01000114 AXCM01000115 AXCM01000116 AXCM01000117 GDIP01104801 GDIQ01050471 JAL98913.1 JAN44266.1 GDIQ01015012 JAN79725.1 GGMS01012516 MBY81719.1 CH963847 EDW72992.2 GDIP01130796 JAL72918.1 NNAY01000465 OXU28180.1 GANO01002411 JAB57460.1 CH902618 EDV40340.1 AAZX01000583 GBXI01008171 JAD06121.1 KK107182 EZA55993.1 JRES01000996 KNC26319.1 CP012525 ALC43144.1 CH916366 EDV96442.1 QOIP01000003 RLU25076.1 QCYY01000412 ROT85188.1 OUUW01000002 SPP76932.1 GECZ01027224 GECZ01004752 GECZ01000645 JAS42545.1 JAS65017.1 JAS69124.1 LBMM01014013 KMQ85380.1 NEVH01013548 PNF28874.1 AJVK01013101 KK857477 PTY27205.1 KK852702 KDR18108.1 GAMC01001927 JAC04629.1 GDAI01002640 JAI14963.1 GAKP01014098 JAC44854.1 CH379069 EAL29482.1 CH479206 EDW30870.1 GL437497 EFN70345.1
KPI93249.1 KQ459937 KPJ19203.1 RSAL01000005 RVE54341.1 JTDY01002339 KOB71636.1 AGBW02007668 OWR54779.1 APGK01047286 KB741077 KB632432 ENN74331.1 ERL95533.1 GAIX01000555 JAA92005.1 GEZM01087797 JAV58471.1 CH477347 EAT42975.1 EJY57594.1 GGMR01002817 MBY15436.1 GAPW01001639 JAC11959.1 APCN01005783 KQ971307 EFA11202.1 GGFJ01005666 MBW54807.1 LJIG01022565 KRT79897.1 GGFK01007955 MBW41276.1 GBGD01001537 JAC87352.1 GGFJ01005665 MBW54806.1 GFXV01001294 MBW13099.1 ADMH02001552 ETN62090.1 ATLV01025286 KE525379 KFB52375.1 JXUM01120885 KQ566440 KXJ70136.1 ABLF02025516 GGFJ01005664 MBW54805.1 GGFM01007382 MBW28133.1 AXCN02001180 AAAB01008898 EAA09142.4 ACPB03018838 GAHY01001790 JAA75720.1 CH940647 EDW69727.2 GBHO01021976 GBRD01005654 GBRD01005652 GBRD01005651 GDHC01000603 JAG21628.1 JAG60167.1 JAQ18026.1 GFDL01008351 JAV26694.1 GECL01001207 JAP04917.1 GFTR01006170 JAW10256.1 DS231912 EDS25837.1 PYGN01000519 PSN44935.1 CM000159 EDW93514.1 CM000363 CM002912 EDX09324.1 KMY97752.1 GDIP01182182 JAJ41220.1 CH954178 EDV51022.1 GL732527 EFX87492.1 CH933809 EDW18354.1 LRGB01002371 KZS08073.1 GDIP01180441 JAJ42961.1 CH480817 EDW50584.1 GDIP01136545 JAL67169.1 GDIP01183637 JAJ39765.1 GBBI01001166 JAC17546.1 AE014296 AY051980 AJ252069 AAF50771.1 AAK93404.1 CAB64384.1 GDIP01152595 JAJ70807.1 GDIP01218478 GDIP01216484 GDIP01204875 JAJ18527.1 AXCM01000114 AXCM01000115 AXCM01000116 AXCM01000117 GDIP01104801 GDIQ01050471 JAL98913.1 JAN44266.1 GDIQ01015012 JAN79725.1 GGMS01012516 MBY81719.1 CH963847 EDW72992.2 GDIP01130796 JAL72918.1 NNAY01000465 OXU28180.1 GANO01002411 JAB57460.1 CH902618 EDV40340.1 AAZX01000583 GBXI01008171 JAD06121.1 KK107182 EZA55993.1 JRES01000996 KNC26319.1 CP012525 ALC43144.1 CH916366 EDV96442.1 QOIP01000003 RLU25076.1 QCYY01000412 ROT85188.1 OUUW01000002 SPP76932.1 GECZ01027224 GECZ01004752 GECZ01000645 JAS42545.1 JAS65017.1 JAS69124.1 LBMM01014013 KMQ85380.1 NEVH01013548 PNF28874.1 AJVK01013101 KK857477 PTY27205.1 KK852702 KDR18108.1 GAMC01001927 JAC04629.1 GDAI01002640 JAI14963.1 GAKP01014098 JAC44854.1 CH379069 EAL29482.1 CH479206 EDW30870.1 GL437497 EFN70345.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000283053
UP000037510
+ More
UP000007151 UP000019118 UP000030742 UP000192223 UP000008820 UP000075880 UP000075882 UP000075881 UP000075903 UP000075902 UP000076407 UP000075840 UP000007266 UP000000673 UP000030765 UP000069940 UP000249989 UP000007819 UP000075886 UP000007062 UP000075885 UP000069272 UP000076408 UP000015103 UP000008792 UP000075920 UP000075884 UP000002320 UP000245037 UP000002282 UP000000304 UP000192221 UP000008711 UP000000305 UP000009192 UP000076858 UP000001292 UP000000803 UP000075883 UP000007798 UP000075900 UP000215335 UP000007801 UP000002358 UP000053097 UP000037069 UP000092553 UP000001070 UP000279307 UP000283509 UP000268350 UP000095301 UP000036403 UP000235965 UP000092462 UP000027135 UP000091820 UP000001819 UP000008744 UP000000311 UP000092443
UP000007151 UP000019118 UP000030742 UP000192223 UP000008820 UP000075880 UP000075882 UP000075881 UP000075903 UP000075902 UP000076407 UP000075840 UP000007266 UP000000673 UP000030765 UP000069940 UP000249989 UP000007819 UP000075886 UP000007062 UP000075885 UP000069272 UP000076408 UP000015103 UP000008792 UP000075920 UP000075884 UP000002320 UP000245037 UP000002282 UP000000304 UP000192221 UP000008711 UP000000305 UP000009192 UP000076858 UP000001292 UP000000803 UP000075883 UP000007798 UP000075900 UP000215335 UP000007801 UP000002358 UP000053097 UP000037069 UP000092553 UP000001070 UP000279307 UP000283509 UP000268350 UP000095301 UP000036403 UP000235965 UP000092462 UP000027135 UP000091820 UP000001819 UP000008744 UP000000311 UP000092443
PRIDE
Interpro
SUPFAM
SSF52210
SSF52210
Gene 3D
ProteinModelPortal
H9JE77
A0A2A4K021
A0A2H1VJW1
A0A194PKJ0
A0A0N0PEI7
A0A3S2P134
+ More
A0A0L7L8M9 A0A212FM66 N6U140 S4PRS6 A0A1Y1KDP3 A0A1W4XAU8 Q179P9 A0A182IRZ2 A0A182LNJ0 A0A182K4E2 A0A2S2NF83 A0A182UQ76 A0A182U3I9 A0A023EU71 A0A182XF24 A0A182HVJ2 D6W7F8 A0A2M4BP07 A0A0T6AXJ2 A0A2M4AKG0 A0A069DUK8 A0A2M4BNY9 A0A2H8TGC3 W5JGQ2 A0A084WQ81 A0A182H9I6 J9K8K5 A0A2M4BP30 A0A2M3ZHZ0 A0A182QGG2 Q7QA95 A0A182P3F3 A0A182FAR5 A0A182YJP7 R4G3Z4 B4LBD9 A0A0A9XS56 A0A1Q3FGM3 A0A0V0GC39 A0A224XCT7 A0A182W339 A0A182NCQ1 B0WEX9 A0A2P8YL05 B4PIU4 B4QRL3 A0A1W4VSI0 A0A0P5BPC8 B3NGC1 E9FY81 B4KVB1 A0A164QUM9 A0A0N8A5I7 B4HUG3 A0A0P5SR39 A0A0P5BL18 A0A023F898 Q9V470 A0A0P5E5E9 A0A0P5A382 A0A182LRP0 A0A0P6GNE8 A0A0N8ELI2 A0A2S2QVC8 B4MLR6 A0A0P5T9D6 A0A182R8H0 A0A232FCG8 U5EJ13 B3M3P7 K7IX86 A0A0A1X5E5 A0A026WJ19 A0A0L0C4E4 A0A0M5IYC5 B4IXS1 A0A3L8DXH5 A0A423U904 A0A3B0JAC1 A0A1I8MY09 A0A1B6H359 A0A0J7MY75 A0A2J7QJW3 A0A1B0GNB0 A0A2R7X8H7 A0A067RDN7 W8C579 A0A0K8TLA4 A0A1A9W8I5 A0A034VQW1 Q2LZM5 B4H3D0 E2A879 A0A1A9XRQ9
A0A0L7L8M9 A0A212FM66 N6U140 S4PRS6 A0A1Y1KDP3 A0A1W4XAU8 Q179P9 A0A182IRZ2 A0A182LNJ0 A0A182K4E2 A0A2S2NF83 A0A182UQ76 A0A182U3I9 A0A023EU71 A0A182XF24 A0A182HVJ2 D6W7F8 A0A2M4BP07 A0A0T6AXJ2 A0A2M4AKG0 A0A069DUK8 A0A2M4BNY9 A0A2H8TGC3 W5JGQ2 A0A084WQ81 A0A182H9I6 J9K8K5 A0A2M4BP30 A0A2M3ZHZ0 A0A182QGG2 Q7QA95 A0A182P3F3 A0A182FAR5 A0A182YJP7 R4G3Z4 B4LBD9 A0A0A9XS56 A0A1Q3FGM3 A0A0V0GC39 A0A224XCT7 A0A182W339 A0A182NCQ1 B0WEX9 A0A2P8YL05 B4PIU4 B4QRL3 A0A1W4VSI0 A0A0P5BPC8 B3NGC1 E9FY81 B4KVB1 A0A164QUM9 A0A0N8A5I7 B4HUG3 A0A0P5SR39 A0A0P5BL18 A0A023F898 Q9V470 A0A0P5E5E9 A0A0P5A382 A0A182LRP0 A0A0P6GNE8 A0A0N8ELI2 A0A2S2QVC8 B4MLR6 A0A0P5T9D6 A0A182R8H0 A0A232FCG8 U5EJ13 B3M3P7 K7IX86 A0A0A1X5E5 A0A026WJ19 A0A0L0C4E4 A0A0M5IYC5 B4IXS1 A0A3L8DXH5 A0A423U904 A0A3B0JAC1 A0A1I8MY09 A0A1B6H359 A0A0J7MY75 A0A2J7QJW3 A0A1B0GNB0 A0A2R7X8H7 A0A067RDN7 W8C579 A0A0K8TLA4 A0A1A9W8I5 A0A034VQW1 Q2LZM5 B4H3D0 E2A879 A0A1A9XRQ9
PDB
5CAE
E-value=1.3538e-119,
Score=1100
Ontologies
PATHWAY
GO
PANTHER
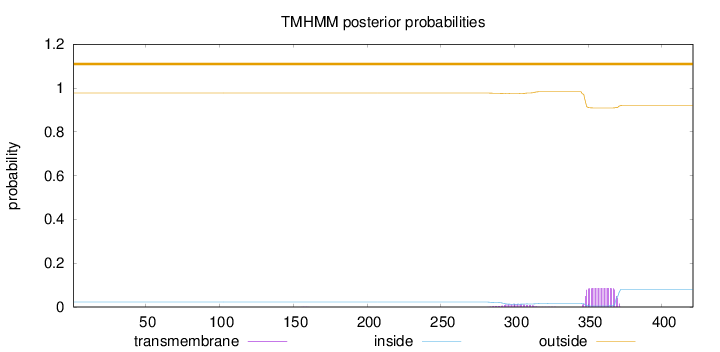
Topology
Subcellular location
Mitochondrion
Length:
421
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.16333
Exp number, first 60 AAs:
0.00186
Total prob of N-in:
0.02298
outside
1 - 421
Population Genetic Test Statistics
Pi
257.983133
Theta
218.873698
Tajima's D
0.666269
CLR
0.68825
CSRT
0.564421778911054
Interpretation
Uncertain
