Gene
KWMTBOMO08891
Annotation
PREDICTED:_NADPH:adrenodoxin_oxidoreductase?_mitochondrial_[Bombyx_mori]
Full name
NADPH:adrenodoxin oxidoreductase, mitochondrial
Alternative Name
Ferredoxin--NADP(+) reductase
Location in the cell
Cytoplasmic Reliability : 1.515 Mitochondrial Reliability : 1.831
Sequence
CDS
ATGACTTTCGGAAATTTAAGAACTTTGAACAGATTTTATTCAGCAAATAATTACACAAAAATTCCTCGAGTATGTATTGTTGGAGCTGGTCCTGCAGGCTTTTATGCCGCCATGCATCTTACGAAAAACATACAATGCAAGATTGATTTAATAGAAAAACTTCCGGTACCCTTCGGATTAATAAGATATGGTGTAGCTCCAGATCACCCTGAAGTAAAAAATGTGATAAATCAATTTACCAAAGTGGCTCAACGACCAGAGGTCAATTTTTATGGAAATGTTACTCTAGGAAAAGATATCACTTTAAATCAACTAAGACAGCATTATGATGCAGTATTATTGACATATGGTGCTGAAAAAGATAAGACTCTTGGTATAGAAAACGAAAACGCCAAGAATGTTATTGGAGCCAGACATTTTGTTGGATGGTATAATGGGCTACCTAGTAATAAAGACCTGGAGATTGATCTATCTTGTGACACGGCTGCTATTCTTGGTCAAGGCAATGTAGCTCTAGATGTTGCAAGAATACTTTTAAGTCCTATTGATGAACTTAGAAAAACTGATATTACAGAACATGCATTAAATATTCTAGCAGGATCAAAAATCAAACAGTTGTATTTGATTGGTCGAAGAGGTCCACTTCAAGTGGCATTCACTATTAAAGAACTTAGAGAACAATTGAAATTAAGAGATTGTAAGACTGTATGGAGGCCAGAAGATTTTCAAAATGTCATCAATGAAGTTCCTAATTTACCACGCCCTAGAAAAAGAATAACTGAACTCATGCTGAAGTCTTTGAACGAAAATGAAACATGTAGCATCTCTAATAATACAAAAATGTTTAAGCCAATTTTTTTCAGAAGCCCAAAAAAATTCATAATTAACGATAAAAATAATGTAACGGGTATAGAGTTGACTTGTAATAAACTTGAAGGTGAAAATTGGGAGAGTCAAAAGTGTATTCCGACTGATGAAATTGAAGTGGTGAATTGTGGTTTAGTTTTCAGAAGTATCGGATACAAAAGTATCAAAGCAGATGAAGATGTGCCATTTAGTAATGGGCTAGTTTTAAATGAAAGAGGTTGTGTTATTGACGAAGCTAATGAGCTAGGAAAACTGTACGTAGCAGGATGGCTCGGTACTGGACCAGTGGGTGTTATTTTGCACACCATGGGAAATGCATTTCAAGTAGCAAAAACGATTTGTGATGATCTGAAAAACAAAGCTGATTATAGTAAAAGTGGCTTTGCTGAATTCAAAGATATAGTAAAAGACAAGACTGATATAGTTGATTGGGAGGGTTGGCAGAAGATAGATAAGTTTGAAATTGAAGAAGGTAAAAAACGTGGAAAACCACGAGAAAAGATTTGTAGTGTAGAAAAAATTATAGAAATAGCTAAATCAGAATTTAACATTGTTAAGGATAATAGTTAA
Protein
MTFGNLRTLNRFYSANNYTKIPRVCIVGAGPAGFYAAMHLTKNIQCKIDLIEKLPVPFGLIRYGVAPDHPEVKNVINQFTKVAQRPEVNFYGNVTLGKDITLNQLRQHYDAVLLTYGAEKDKTLGIENENAKNVIGARHFVGWYNGLPSNKDLEIDLSCDTAAILGQGNVALDVARILLSPIDELRKTDITEHALNILAGSKIKQLYLIGRRGPLQVAFTIKELREQLKLRDCKTVWRPEDFQNVINEVPNLPRPRKRITELMLKSLNENETCSISNNTKMFKPIFFRSPKKFIINDKNNVTGIELTCNKLEGENWESQKCIPTDEIEVVNCGLVFRSIGYKSIKADEDVPFSNGLVLNERGCVIDEANELGKLYVAGWLGTGPVGVILHTMGNAFQVAKTICDDLKNKADYSKSGFAEFKDIVKDKTDIVDWEGWQKIDKFEIEEGKKRGKPREKICSVEKIIEIAKSEFNIVKDNS
Summary
Description
Required for synthesis of steroid hormones, for olfactory sensory behavior and completion of the second larval molt (a steroid mediated developmental transition) and pupariation.
Catalytic Activity
H(+) + NADP(+) + 2 reduced [adrenodoxin] = NADPH + 2 oxidized [adrenodoxin]
Cofactor
FAD
Similarity
Belongs to the ferredoxin--NADP reductase type 1 family.
Belongs to the universal ribosomal protein uS7 family.
Belongs to the universal ribosomal protein uS7 family.
Keywords
Complete proteome
Electron transport
FAD
Flavoprotein
Membrane
Mitochondrion
Mitochondrion inner membrane
NADP
Oxidoreductase
Reference proteome
Transit peptide
Transport
Feature
chain NADPH:adrenodoxin oxidoreductase, mitochondrial
Uniprot
A0A2A4K1H7
A0A2H1VJW3
A0A0N1PHC9
A0A194PIV5
A0A0L7LR66
A0A212FKT7
+ More
A0A0A1XCN3 A0A034W9E0 A0A1A9V0R4 A0A1B0FER1 A0A1A9X3I9 A0A0L0BPB6 A0A3L8E4L9 A0A1A9ZUR3 A0A1I8P8K2 A0A3B0JU50 A0A182MDM5 Q174X5 A0A182QS53 A0A1J1IJV3 A0A1Q3FXU0 A0A1I8MW12 B4MPD7 Q563C8 A0A0L7QLB4 W8ALN6 E2B776 Q28YX8 A0A1B6CMT6 A0A1Y1MNM1 B4KRQ8 A0A1B6KKK1 A0A2M4AMK4 A0A182WLT9 A0A182JV19 A0A182RC28 A0A182NMD3 B4MFM6 A0A084WDB8 A0A1B6FWF6 A0A2J7PXF3 A0A182ISJ6 A0A2M4AMN6 A0A1B6HYA3 A0A023ET68 A0A182GH14 A0A026WD51 W5JN26 A0A182PVJ2 A0A182GH15 A0A182UV01 A0A182HM91 A0A0N0BK36 Q7Q7A2 A0A182Y0R3 A0A182FHP2 Q4U479 B4J8H1 A0A182LJU6 A0A195DZQ4 A0A0K8U3A3 A0A182XCW2 A0A158NER3 A0A195AVM9 A0A0M5IXH9 B4GIJ4 B4QB68 A0A195FM02 C4IY01 Q9V3T9 Q8IGW0 A0A195CRB1 A0A067QIJ6 B4P7H3 B3NSJ6 B4HNE3 A0A1B0GLQ9 V3ZX23 F4WRN3 C3YC80 A0A3Q2YFL0 A0A1W4U5L2 A0A232EI20 E2A158 B3MEU2 E9IAA2 H3ASY8 A0A091TCF0 E6ZGI4 K7FK23 A0A2P4TGP2 A0A093QHD8 A0A1V4KVV0 A0A1W4WWI9 A7RUM6 A0A3Q0G0I8 A0A1D5NXA0 A0A3B3DC21 A0A3B4U2E5 M7B1I9
A0A0A1XCN3 A0A034W9E0 A0A1A9V0R4 A0A1B0FER1 A0A1A9X3I9 A0A0L0BPB6 A0A3L8E4L9 A0A1A9ZUR3 A0A1I8P8K2 A0A3B0JU50 A0A182MDM5 Q174X5 A0A182QS53 A0A1J1IJV3 A0A1Q3FXU0 A0A1I8MW12 B4MPD7 Q563C8 A0A0L7QLB4 W8ALN6 E2B776 Q28YX8 A0A1B6CMT6 A0A1Y1MNM1 B4KRQ8 A0A1B6KKK1 A0A2M4AMK4 A0A182WLT9 A0A182JV19 A0A182RC28 A0A182NMD3 B4MFM6 A0A084WDB8 A0A1B6FWF6 A0A2J7PXF3 A0A182ISJ6 A0A2M4AMN6 A0A1B6HYA3 A0A023ET68 A0A182GH14 A0A026WD51 W5JN26 A0A182PVJ2 A0A182GH15 A0A182UV01 A0A182HM91 A0A0N0BK36 Q7Q7A2 A0A182Y0R3 A0A182FHP2 Q4U479 B4J8H1 A0A182LJU6 A0A195DZQ4 A0A0K8U3A3 A0A182XCW2 A0A158NER3 A0A195AVM9 A0A0M5IXH9 B4GIJ4 B4QB68 A0A195FM02 C4IY01 Q9V3T9 Q8IGW0 A0A195CRB1 A0A067QIJ6 B4P7H3 B3NSJ6 B4HNE3 A0A1B0GLQ9 V3ZX23 F4WRN3 C3YC80 A0A3Q2YFL0 A0A1W4U5L2 A0A232EI20 E2A158 B3MEU2 E9IAA2 H3ASY8 A0A091TCF0 E6ZGI4 K7FK23 A0A2P4TGP2 A0A093QHD8 A0A1V4KVV0 A0A1W4WWI9 A7RUM6 A0A3Q0G0I8 A0A1D5NXA0 A0A3B3DC21 A0A3B4U2E5 M7B1I9
EC Number
1.18.1.6
Pubmed
26354079
26227816
22118469
25830018
25348373
26108605
+ More
30249741 17510324 25315136 17994087 15804580 24495485 20798317 15632085 28004739 24438588 24945155 26483478 24508170 20920257 23761445 12364791 14747013 17210077 25244985 20966253 21347285 22936249 10498693 10731132 12537572 24845553 17550304 23254933 21719571 18563158 28648823 21282665 9215903 17381049 17615350 15592404 29451363 23624526
30249741 17510324 25315136 17994087 15804580 24495485 20798317 15632085 28004739 24438588 24945155 26483478 24508170 20920257 23761445 12364791 14747013 17210077 25244985 20966253 21347285 22936249 10498693 10731132 12537572 24845553 17550304 23254933 21719571 18563158 28648823 21282665 9215903 17381049 17615350 15592404 29451363 23624526
EMBL
NWSH01000302
PCG77613.1
ODYU01002957
SOQ41115.1
KQ459937
KPJ19209.1
+ More
KQ459602 KPI93252.1 JTDY01000275 KOB77952.1 AGBW02007963 OWR54361.1 GBXI01005580 JAD08712.1 GAKP01008222 JAC50730.1 CCAG010009035 JRES01001578 KNC21828.1 QOIP01000001 RLU27365.1 OUUW01000001 SPP74598.1 AXCM01007726 CH477404 EAT41657.1 AXCN02000934 CVRI01000054 CRL00472.1 GFDL01002753 JAV32292.1 CH963849 EDW73976.1 AY947548 AAX85204.1 KQ414924 KOC59395.1 GAMC01016905 JAB89650.1 GL446149 EFN88437.1 CM000071 EAL25838.1 GEDC01022514 JAS14784.1 GEZM01026070 JAV87261.1 CH933808 EDW08328.1 GEBQ01027998 JAT11979.1 GGFK01008688 MBW42009.1 CH940667 EDW57197.2 ATLV01022985 KE525339 KFB48212.1 GECZ01015318 JAS54451.1 NEVH01020859 PNF20994.1 GGFK01008719 MBW42040.1 GECU01037318 GECU01028057 JAS70388.1 JAS79649.1 GAPW01001115 JAC12483.1 JXUM01062870 KQ562219 KXJ76369.1 KK107260 EZA54000.1 ADMH02001054 ETN64295.1 KXJ76370.1 APCN01004380 KQ435709 KOX80011.1 AAAB01008960 EAA11924.4 DQ013245 AAY34441.1 CH916367 EDW02330.1 KQ979999 KYN18311.1 GDHF01031127 GDHF01006350 JAI21187.1 JAI45964.1 ADTU01013612 KQ976736 KYM76117.1 CP012524 ALC41486.1 CH479183 EDW36314.1 CM000362 CM002911 EDX06601.1 KMY92958.1 KQ981490 KYN41282.1 BT083432 ACQ99837.1 AF168685 AE013599 BT001566 AAN71321.1 KQ977381 KYN03175.1 KK853521 KDR07014.1 CM000158 EDW90008.1 CH954179 EDV56498.1 CH480816 EDW47379.1 AJWK01032941 KB202953 ESO87175.1 GL888292 EGI63040.1 GG666500 EEN62112.1 NNAY01004368 OXU17995.1 GL435707 EFN72825.1 CH902619 EDV35556.1 GL761997 EFZ22498.1 AFYH01091804 AFYH01091805 KK469277 KFQ56531.1 FQ310507 CBN81168.1 AGCU01174015 AGCU01174016 AGCU01174017 AGCU01174018 PPHD01000408 POI35519.1 KL671685 KFW83482.1 LSYS01001520 OPJ88629.1 DS469540 EDO44879.1 AADN05000628 KB547685 EMP30904.1
KQ459602 KPI93252.1 JTDY01000275 KOB77952.1 AGBW02007963 OWR54361.1 GBXI01005580 JAD08712.1 GAKP01008222 JAC50730.1 CCAG010009035 JRES01001578 KNC21828.1 QOIP01000001 RLU27365.1 OUUW01000001 SPP74598.1 AXCM01007726 CH477404 EAT41657.1 AXCN02000934 CVRI01000054 CRL00472.1 GFDL01002753 JAV32292.1 CH963849 EDW73976.1 AY947548 AAX85204.1 KQ414924 KOC59395.1 GAMC01016905 JAB89650.1 GL446149 EFN88437.1 CM000071 EAL25838.1 GEDC01022514 JAS14784.1 GEZM01026070 JAV87261.1 CH933808 EDW08328.1 GEBQ01027998 JAT11979.1 GGFK01008688 MBW42009.1 CH940667 EDW57197.2 ATLV01022985 KE525339 KFB48212.1 GECZ01015318 JAS54451.1 NEVH01020859 PNF20994.1 GGFK01008719 MBW42040.1 GECU01037318 GECU01028057 JAS70388.1 JAS79649.1 GAPW01001115 JAC12483.1 JXUM01062870 KQ562219 KXJ76369.1 KK107260 EZA54000.1 ADMH02001054 ETN64295.1 KXJ76370.1 APCN01004380 KQ435709 KOX80011.1 AAAB01008960 EAA11924.4 DQ013245 AAY34441.1 CH916367 EDW02330.1 KQ979999 KYN18311.1 GDHF01031127 GDHF01006350 JAI21187.1 JAI45964.1 ADTU01013612 KQ976736 KYM76117.1 CP012524 ALC41486.1 CH479183 EDW36314.1 CM000362 CM002911 EDX06601.1 KMY92958.1 KQ981490 KYN41282.1 BT083432 ACQ99837.1 AF168685 AE013599 BT001566 AAN71321.1 KQ977381 KYN03175.1 KK853521 KDR07014.1 CM000158 EDW90008.1 CH954179 EDV56498.1 CH480816 EDW47379.1 AJWK01032941 KB202953 ESO87175.1 GL888292 EGI63040.1 GG666500 EEN62112.1 NNAY01004368 OXU17995.1 GL435707 EFN72825.1 CH902619 EDV35556.1 GL761997 EFZ22498.1 AFYH01091804 AFYH01091805 KK469277 KFQ56531.1 FQ310507 CBN81168.1 AGCU01174015 AGCU01174016 AGCU01174017 AGCU01174018 PPHD01000408 POI35519.1 KL671685 KFW83482.1 LSYS01001520 OPJ88629.1 DS469540 EDO44879.1 AADN05000628 KB547685 EMP30904.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000037510
UP000007151
UP000078200
+ More
UP000092444 UP000091820 UP000037069 UP000279307 UP000092445 UP000095300 UP000268350 UP000075883 UP000008820 UP000075886 UP000183832 UP000095301 UP000007798 UP000053825 UP000008237 UP000001819 UP000009192 UP000075920 UP000075881 UP000075900 UP000075884 UP000008792 UP000030765 UP000235965 UP000075880 UP000069940 UP000249989 UP000053097 UP000000673 UP000075885 UP000075903 UP000075840 UP000053105 UP000007062 UP000076408 UP000069272 UP000001070 UP000075882 UP000078492 UP000076407 UP000005205 UP000078540 UP000092553 UP000008744 UP000000304 UP000078541 UP000000803 UP000078542 UP000027135 UP000002282 UP000008711 UP000001292 UP000092461 UP000030746 UP000007755 UP000001554 UP000264820 UP000192221 UP000215335 UP000000311 UP000007801 UP000008672 UP000007267 UP000053258 UP000190648 UP000192223 UP000001593 UP000189705 UP000000539 UP000261560 UP000261420 UP000031443
UP000092444 UP000091820 UP000037069 UP000279307 UP000092445 UP000095300 UP000268350 UP000075883 UP000008820 UP000075886 UP000183832 UP000095301 UP000007798 UP000053825 UP000008237 UP000001819 UP000009192 UP000075920 UP000075881 UP000075900 UP000075884 UP000008792 UP000030765 UP000235965 UP000075880 UP000069940 UP000249989 UP000053097 UP000000673 UP000075885 UP000075903 UP000075840 UP000053105 UP000007062 UP000076408 UP000069272 UP000001070 UP000075882 UP000078492 UP000076407 UP000005205 UP000078540 UP000092553 UP000008744 UP000000304 UP000078541 UP000000803 UP000078542 UP000027135 UP000002282 UP000008711 UP000001292 UP000092461 UP000030746 UP000007755 UP000001554 UP000264820 UP000192221 UP000215335 UP000000311 UP000007801 UP000008672 UP000007267 UP000053258 UP000190648 UP000192223 UP000001593 UP000189705 UP000000539 UP000261560 UP000261420 UP000031443
Interpro
IPR021163
Ferredox_Rdtase_adrenod
+ More
IPR036188 FAD/NAD-bd_sf
IPR023753 FAD/NAD-binding_dom
IPR032394 Anoct_dimer
IPR036823 Ribosomal_S7_dom_sf
IPR020606 Ribosomal_S7_CS
IPR005716 Ribosomal_S5/S7_euk/arc
IPR000235 Ribosomal_S5/S7
IPR023798 Ribosomal_S7_dom
IPR029063 SAM-dependent_MTases
IPR025807 Adrift-typ_MeTrfase
IPR002877 rRNA_MeTrfase_FtsJ_dom
IPR007726 SS18_fam
IPR036188 FAD/NAD-bd_sf
IPR023753 FAD/NAD-binding_dom
IPR032394 Anoct_dimer
IPR036823 Ribosomal_S7_dom_sf
IPR020606 Ribosomal_S7_CS
IPR005716 Ribosomal_S5/S7_euk/arc
IPR000235 Ribosomal_S5/S7
IPR023798 Ribosomal_S7_dom
IPR029063 SAM-dependent_MTases
IPR025807 Adrift-typ_MeTrfase
IPR002877 rRNA_MeTrfase_FtsJ_dom
IPR007726 SS18_fam
Gene 3D
ProteinModelPortal
A0A2A4K1H7
A0A2H1VJW3
A0A0N1PHC9
A0A194PIV5
A0A0L7LR66
A0A212FKT7
+ More
A0A0A1XCN3 A0A034W9E0 A0A1A9V0R4 A0A1B0FER1 A0A1A9X3I9 A0A0L0BPB6 A0A3L8E4L9 A0A1A9ZUR3 A0A1I8P8K2 A0A3B0JU50 A0A182MDM5 Q174X5 A0A182QS53 A0A1J1IJV3 A0A1Q3FXU0 A0A1I8MW12 B4MPD7 Q563C8 A0A0L7QLB4 W8ALN6 E2B776 Q28YX8 A0A1B6CMT6 A0A1Y1MNM1 B4KRQ8 A0A1B6KKK1 A0A2M4AMK4 A0A182WLT9 A0A182JV19 A0A182RC28 A0A182NMD3 B4MFM6 A0A084WDB8 A0A1B6FWF6 A0A2J7PXF3 A0A182ISJ6 A0A2M4AMN6 A0A1B6HYA3 A0A023ET68 A0A182GH14 A0A026WD51 W5JN26 A0A182PVJ2 A0A182GH15 A0A182UV01 A0A182HM91 A0A0N0BK36 Q7Q7A2 A0A182Y0R3 A0A182FHP2 Q4U479 B4J8H1 A0A182LJU6 A0A195DZQ4 A0A0K8U3A3 A0A182XCW2 A0A158NER3 A0A195AVM9 A0A0M5IXH9 B4GIJ4 B4QB68 A0A195FM02 C4IY01 Q9V3T9 Q8IGW0 A0A195CRB1 A0A067QIJ6 B4P7H3 B3NSJ6 B4HNE3 A0A1B0GLQ9 V3ZX23 F4WRN3 C3YC80 A0A3Q2YFL0 A0A1W4U5L2 A0A232EI20 E2A158 B3MEU2 E9IAA2 H3ASY8 A0A091TCF0 E6ZGI4 K7FK23 A0A2P4TGP2 A0A093QHD8 A0A1V4KVV0 A0A1W4WWI9 A7RUM6 A0A3Q0G0I8 A0A1D5NXA0 A0A3B3DC21 A0A3B4U2E5 M7B1I9
A0A0A1XCN3 A0A034W9E0 A0A1A9V0R4 A0A1B0FER1 A0A1A9X3I9 A0A0L0BPB6 A0A3L8E4L9 A0A1A9ZUR3 A0A1I8P8K2 A0A3B0JU50 A0A182MDM5 Q174X5 A0A182QS53 A0A1J1IJV3 A0A1Q3FXU0 A0A1I8MW12 B4MPD7 Q563C8 A0A0L7QLB4 W8ALN6 E2B776 Q28YX8 A0A1B6CMT6 A0A1Y1MNM1 B4KRQ8 A0A1B6KKK1 A0A2M4AMK4 A0A182WLT9 A0A182JV19 A0A182RC28 A0A182NMD3 B4MFM6 A0A084WDB8 A0A1B6FWF6 A0A2J7PXF3 A0A182ISJ6 A0A2M4AMN6 A0A1B6HYA3 A0A023ET68 A0A182GH14 A0A026WD51 W5JN26 A0A182PVJ2 A0A182GH15 A0A182UV01 A0A182HM91 A0A0N0BK36 Q7Q7A2 A0A182Y0R3 A0A182FHP2 Q4U479 B4J8H1 A0A182LJU6 A0A195DZQ4 A0A0K8U3A3 A0A182XCW2 A0A158NER3 A0A195AVM9 A0A0M5IXH9 B4GIJ4 B4QB68 A0A195FM02 C4IY01 Q9V3T9 Q8IGW0 A0A195CRB1 A0A067QIJ6 B4P7H3 B3NSJ6 B4HNE3 A0A1B0GLQ9 V3ZX23 F4WRN3 C3YC80 A0A3Q2YFL0 A0A1W4U5L2 A0A232EI20 E2A158 B3MEU2 E9IAA2 H3ASY8 A0A091TCF0 E6ZGI4 K7FK23 A0A2P4TGP2 A0A093QHD8 A0A1V4KVV0 A0A1W4WWI9 A7RUM6 A0A3Q0G0I8 A0A1D5NXA0 A0A3B3DC21 A0A3B4U2E5 M7B1I9
PDB
1E6E
E-value=7.71476e-104,
Score=964
Ontologies
GO
GO:0015039
GO:0005739
GO:0046983
GO:0005840
GO:0003735
GO:0006412
GO:0003723
GO:0015935
GO:0042048
GO:0035073
GO:0007591
GO:0007552
GO:0006694
GO:0008168
GO:0016491
GO:0006744
GO:0003713
GO:0006707
GO:0009055
GO:0005759
GO:0005743
GO:0006886
GO:0016192
GO:0030131
GO:0016020
GO:0016817
GO:0008270
GO:0043565
GO:0016876
GO:0043039
GO:0055114
PANTHER
Topology
Subcellular location
Mitochondrion
Mitochondrion inner membrane
Mitochondrion inner membrane
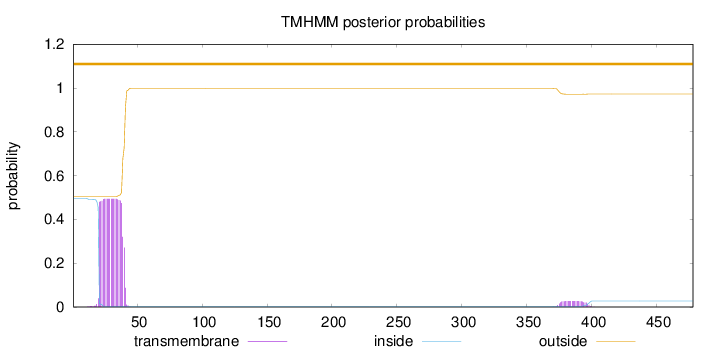
Length:
478
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
10.18481
Exp number, first 60 AAs:
9.59964999999999
Total prob of N-in:
0.49501
outside
1 - 478
Population Genetic Test Statistics
Pi
269.555344
Theta
199.661761
Tajima's D
1.101864
CLR
0
CSRT
0.690665466726664
Interpretation
Uncertain
