Pre Gene Modal
BGIBMGA007825
Annotation
PREDICTED:_AP-3_complex_subunit_mu-1_[Amyelois_transitella]
Location in the cell
Golgi Reliability : 1.121 PlasmaMembrane Reliability : 1.101
Sequence
CDS
ATGATACACAGTTTATTTATAATAAATCCATCGGGCGACGTGTTTCTAGAGAAGCATTGGAGAAGTGTGATCCCTCGTTCAGTATGTGATTATTATTTGGAAGCACAGAGAGCATCACCTAATGATGTACCACCGGTGTTGGCTGCTCCACATCATTACCTCATATCAATTCATAGAGGAGGAGTTGCACTTGTTGCTGTCTGTAAGCAGGAAGTTGCCCCACTATTTGTTATAGAGTTCTTGCATAGAGTTGTTGATACTTTTCAGGACTACTTTTCAGATTGCACTGAGACAATAATTAAAGAAAATTATGTGGTGGTGTATGAATTATTGGATGAAATGTTAGATAATGGCTTCCCACTGGCCACGGAAAGTAATATATTGAAAGAATTGATTAAACCACCAAATATTTTAAGAACAATTGCCAATACTGTTACAGGAAAATCAAATGTATCCTCTACACTTCCGTCTGGACAGCTGTCAAATGTACCTTGGAGAAGGTCAGGTGTAAAGTATGCTAACAATGAAGCTTACTTTGATGTAGTTGAAGAAGTCGATGCAATTATTGACAAAAGTGGAGCTACCGTAAATGCAGAAATACAAGGCTATATTGATTGCTGTATAAAACTGAGCGGAATGCCAGATCTTACTCTCACTTTTGTAAATCCAAGATTGTTTGATGATGTTTCATTTCATCCGTGTGTGCGGTTTAAGCGTTGGGAGGCGGAACGTATTTTATCGTTCATACCGCCCGATGGTAACTTCCGGTTGATGTCGTACCATATAGGATCACAAAGCGTTGTGGCTATACCTATATACGTACGTCATAGCTTGACATTGAGGTCGAATGGGGATCAGGGACGATTAGACCTCACTGTTGGACCAAAGCAGACCATGGGGAGGACGTTAGAAAATGTTGCATTGGAGATTTGCATGCCGAAGTGTGTTCTAAACTGCTGTCTAACAGCGAATCAAGGGAAATATTCCTATGACCCGGTCAGCAAGATGCTGCTTTGGGATATAGGACGTATAGAACTACCCAAACTGCCTAATATTAAAGGGAGCGTGTCGGTGGTATCGGGAGCTGATACGACGGGCGCCAGTCCGAGCATCAACGTTCATTTCACGATTCCGCAATTGGCGGTCAGCGGTCTCAGAGTGAGCCGCCTGGACATGTACGGCGCCAAGTACAAGCCATTCAAAGGCGTCAAGTACGTCACGAAAGCGGGCAAGTTCCACGTCAGGATGTGA
Protein
MIHSLFIINPSGDVFLEKHWRSVIPRSVCDYYLEAQRASPNDVPPVLAAPHHYLISIHRGGVALVAVCKQEVAPLFVIEFLHRVVDTFQDYFSDCTETIIKENYVVVYELLDEMLDNGFPLATESNILKELIKPPNILRTIANTVTGKSNVSSTLPSGQLSNVPWRRSGVKYANNEAYFDVVEEVDAIIDKSGATVNAEIQGYIDCCIKLSGMPDLTLTFVNPRLFDDVSFHPCVRFKRWEAERILSFIPPDGNFRLMSYHIGSQSVVAIPIYVRHSLTLRSNGDQGRLDLTVGPKQTMGRTLENVALEICMPKCVLNCCLTANQGKYSYDPVSKMLLWDIGRIELPKLPNIKGSVSVVSGADTTGASPSINVHFTIPQLAVSGLRVSRLDMYGAKYKPFKGVKYVTKAGKFHVRM
Summary
Similarity
Belongs to the adaptor complexes medium subunit family.
Uniprot
H9JE78
A0A3S2LI51
A0A2H1W3L6
A0A2A4K0M3
A0A194PIV0
A0A0N1PJR3
+ More
A0A1E1WKG0 A0A212FKU9 A0A1Y1LXZ9 D6X4J2 A0A1W4XGA9 N6SWJ6 A0A1L8DWE5 A0A1L8DVV8 W8BVN0 A0A2A3EB91 A0A088A2H6 U5EXH0 A0A0M5J2Y4 A0A0A1WQU2 A0A026WEC6 A0A034VAK3 A0A310SW74 B4JMT2 A0A0J7L0I5 A0A0K8VW77 A0A154NY20 A0A336MTI7 A0A3B0K3M6 B4NEG2 B4L566 Q29GY7 E2BA54 A0A1W4UW73 A0A1I8NYC1 B3MQ12 E9IFD1 B4M9W1 A0A0L0BU93 E2A3P5 A0A0T6B4N0 A0A1I8M1E4 B4IHI7 A0A151IJS6 A0A151JB39 F4WWC5 A0A195B073 A0A195FCM0 A0A158NKJ6 A0A151WRP9 O76928 B4Q159 A0A1A9XRV6 A0A1B0AP73 B3NXY4 A0A1A9ZE61 A0A1A9UUP0 A0A336LZ39 A0A1S4FTJ5 A0A2J7R1C3 A0A1B6K793 Q0IEC0 A0A0N1IT56 A0A1B6GN75 A0A182GZT4 A0A182IMQ8 A0A1B0FQM7 A0A182V0N7 A0A0A9WH95 A0A1B6KHT4 A0A224XD99 A0A023F5N5 A0A182HL04 A0A069DU23 A0A182XA25 A0A182PH03 A0A182UJQ2 A0A1B6CS50 A0A182QEN2 A0A336MWC8 A0A2M3Z9D8 A0A182FRS3 A0A2M4BPW4 A0A0V0G7J8 A0A2M4ANP9 Q7PXM8 A0A423SW68 A0A2M3Z934 W5JHX4 A0A182MJC8 A0A182YB98 A0A0P4VTB8 A0A182R514 A0A182VWD3 A0A182L9C4 J9JYV0 A0A1D2N4A9 A0A182N547 A0A336LZ45 A0A2H8U036
A0A1E1WKG0 A0A212FKU9 A0A1Y1LXZ9 D6X4J2 A0A1W4XGA9 N6SWJ6 A0A1L8DWE5 A0A1L8DVV8 W8BVN0 A0A2A3EB91 A0A088A2H6 U5EXH0 A0A0M5J2Y4 A0A0A1WQU2 A0A026WEC6 A0A034VAK3 A0A310SW74 B4JMT2 A0A0J7L0I5 A0A0K8VW77 A0A154NY20 A0A336MTI7 A0A3B0K3M6 B4NEG2 B4L566 Q29GY7 E2BA54 A0A1W4UW73 A0A1I8NYC1 B3MQ12 E9IFD1 B4M9W1 A0A0L0BU93 E2A3P5 A0A0T6B4N0 A0A1I8M1E4 B4IHI7 A0A151IJS6 A0A151JB39 F4WWC5 A0A195B073 A0A195FCM0 A0A158NKJ6 A0A151WRP9 O76928 B4Q159 A0A1A9XRV6 A0A1B0AP73 B3NXY4 A0A1A9ZE61 A0A1A9UUP0 A0A336LZ39 A0A1S4FTJ5 A0A2J7R1C3 A0A1B6K793 Q0IEC0 A0A0N1IT56 A0A1B6GN75 A0A182GZT4 A0A182IMQ8 A0A1B0FQM7 A0A182V0N7 A0A0A9WH95 A0A1B6KHT4 A0A224XD99 A0A023F5N5 A0A182HL04 A0A069DU23 A0A182XA25 A0A182PH03 A0A182UJQ2 A0A1B6CS50 A0A182QEN2 A0A336MWC8 A0A2M3Z9D8 A0A182FRS3 A0A2M4BPW4 A0A0V0G7J8 A0A2M4ANP9 Q7PXM8 A0A423SW68 A0A2M3Z934 W5JHX4 A0A182MJC8 A0A182YB98 A0A0P4VTB8 A0A182R514 A0A182VWD3 A0A182L9C4 J9JYV0 A0A1D2N4A9 A0A182N547 A0A336LZ45 A0A2H8U036
Pubmed
19121390
26354079
22118469
28004739
18362917
19820115
+ More
23537049 24495485 25830018 24508170 30249741 25348373 17994087 15632085 20798317 21282665 26108605 25315136 21719571 21347285 10375633 10589826 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 17510324 26483478 25401762 26823975 25474469 26334808 12364791 14747013 17210077 20920257 23761445 25244985 27129103 20966253 27289101
23537049 24495485 25830018 24508170 30249741 25348373 17994087 15632085 20798317 21282665 26108605 25315136 21719571 21347285 10375633 10589826 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 17510324 26483478 25401762 26823975 25474469 26334808 12364791 14747013 17210077 20920257 23761445 25244985 27129103 20966253 27289101
EMBL
BABH01002723
RSAL01000005
RVE54342.1
ODYU01005980
SOQ47422.1
NWSH01000302
+ More
PCG77609.1 KQ459602 KPI93247.1 KQ459937 KPJ19201.1 GDQN01003544 JAT87510.1 AGBW02007963 OWR54362.1 GEZM01044221 JAV78379.1 KQ971380 EEZ97259.1 APGK01053948 KB741239 KB632275 ENN72149.1 ERL91121.1 GFDF01003509 JAV10575.1 GFDF01003510 JAV10574.1 GAMC01009199 GAMC01009197 JAB97356.1 KZ288311 PBC28456.1 GANO01000955 JAB58916.1 CP012528 ALC48261.1 GBXI01012873 JAD01419.1 KK107249 QOIP01000013 EZA54455.1 RLU15512.1 GAKP01020157 JAC38795.1 KQ759843 OAD62593.1 CH916371 EDV92025.1 LBMM01001444 KMQ96307.1 GDHF01009176 JAI43138.1 KQ434772 KZC03918.1 UFQS01002419 UFQT01002419 SSX14001.1 SSX33420.1 OUUW01000016 SPP88804.1 CH964239 EDW82131.1 CH933811 EDW06325.1 CH379064 EAL31972.1 GL446664 EFN87463.1 CH902621 EDV44438.1 GL762838 EFZ20694.1 CH940655 EDW66020.1 JRES01001325 KNC23617.1 GL436457 EFN71937.1 LJIG01009955 KRT82045.1 CH480839 EDW49338.1 KQ977294 KYN03897.1 KQ979182 KYN22324.1 GL888406 EGI61467.1 KQ976692 KYM77878.1 KQ981693 KYN37789.1 ADTU01018869 KQ982805 KYQ50483.1 AF110233 AE014298 AY069388 AJ009657 AAF14249.1 AAF46231.1 AAL39533.1 AGB95147.1 CAA08768.1 CM000162 EDX02414.1 JXJN01001236 CH954180 EDV47435.1 UFQT01000325 SSX23255.1 NEVH01008208 PNF34639.1 GECU01000386 JAT07321.1 CH477764 EAT36491.1 KQ435881 KOX69821.1 GECZ01005884 JAS63885.1 JXUM01002161 CCAG010017999 GBHO01035782 GDHC01006737 JAG07822.1 JAQ11892.1 GEBQ01028957 JAT11020.1 GFTR01006011 JAW10415.1 GBBI01002235 JAC16477.1 APCN01000876 GBGD01001623 JAC87266.1 GEDC01021101 JAS16197.1 AXCN02001521 SSX14000.1 SSX33419.1 GGFM01004297 MBW25048.1 GGFJ01005820 MBW54961.1 GECL01002025 JAP04099.1 GGFK01009093 MBW42414.1 AAAB01008987 EAA00857.4 QCYY01002671 ROT68411.1 GGFM01004296 MBW25047.1 ADMH02001137 ETN63967.1 AXCM01004179 GDKW01000744 JAI55851.1 ABLF02025906 LJIJ01000256 ODM99765.1 SSX23254.1 GFXV01008130 MBW19935.1
PCG77609.1 KQ459602 KPI93247.1 KQ459937 KPJ19201.1 GDQN01003544 JAT87510.1 AGBW02007963 OWR54362.1 GEZM01044221 JAV78379.1 KQ971380 EEZ97259.1 APGK01053948 KB741239 KB632275 ENN72149.1 ERL91121.1 GFDF01003509 JAV10575.1 GFDF01003510 JAV10574.1 GAMC01009199 GAMC01009197 JAB97356.1 KZ288311 PBC28456.1 GANO01000955 JAB58916.1 CP012528 ALC48261.1 GBXI01012873 JAD01419.1 KK107249 QOIP01000013 EZA54455.1 RLU15512.1 GAKP01020157 JAC38795.1 KQ759843 OAD62593.1 CH916371 EDV92025.1 LBMM01001444 KMQ96307.1 GDHF01009176 JAI43138.1 KQ434772 KZC03918.1 UFQS01002419 UFQT01002419 SSX14001.1 SSX33420.1 OUUW01000016 SPP88804.1 CH964239 EDW82131.1 CH933811 EDW06325.1 CH379064 EAL31972.1 GL446664 EFN87463.1 CH902621 EDV44438.1 GL762838 EFZ20694.1 CH940655 EDW66020.1 JRES01001325 KNC23617.1 GL436457 EFN71937.1 LJIG01009955 KRT82045.1 CH480839 EDW49338.1 KQ977294 KYN03897.1 KQ979182 KYN22324.1 GL888406 EGI61467.1 KQ976692 KYM77878.1 KQ981693 KYN37789.1 ADTU01018869 KQ982805 KYQ50483.1 AF110233 AE014298 AY069388 AJ009657 AAF14249.1 AAF46231.1 AAL39533.1 AGB95147.1 CAA08768.1 CM000162 EDX02414.1 JXJN01001236 CH954180 EDV47435.1 UFQT01000325 SSX23255.1 NEVH01008208 PNF34639.1 GECU01000386 JAT07321.1 CH477764 EAT36491.1 KQ435881 KOX69821.1 GECZ01005884 JAS63885.1 JXUM01002161 CCAG010017999 GBHO01035782 GDHC01006737 JAG07822.1 JAQ11892.1 GEBQ01028957 JAT11020.1 GFTR01006011 JAW10415.1 GBBI01002235 JAC16477.1 APCN01000876 GBGD01001623 JAC87266.1 GEDC01021101 JAS16197.1 AXCN02001521 SSX14000.1 SSX33419.1 GGFM01004297 MBW25048.1 GGFJ01005820 MBW54961.1 GECL01002025 JAP04099.1 GGFK01009093 MBW42414.1 AAAB01008987 EAA00857.4 QCYY01002671 ROT68411.1 GGFM01004296 MBW25047.1 ADMH02001137 ETN63967.1 AXCM01004179 GDKW01000744 JAI55851.1 ABLF02025906 LJIJ01000256 ODM99765.1 SSX23254.1 GFXV01008130 MBW19935.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000053240
UP000007151
+ More
UP000007266 UP000192223 UP000019118 UP000030742 UP000242457 UP000005203 UP000092553 UP000053097 UP000279307 UP000001070 UP000036403 UP000076502 UP000268350 UP000007798 UP000009192 UP000001819 UP000008237 UP000192221 UP000095300 UP000007801 UP000008792 UP000037069 UP000000311 UP000095301 UP000001292 UP000078542 UP000078492 UP000007755 UP000078540 UP000078541 UP000005205 UP000075809 UP000000803 UP000002282 UP000092443 UP000092460 UP000008711 UP000092445 UP000078200 UP000235965 UP000008820 UP000053105 UP000069940 UP000075880 UP000092444 UP000075903 UP000075840 UP000076407 UP000075885 UP000075902 UP000075886 UP000069272 UP000007062 UP000283509 UP000000673 UP000075883 UP000076408 UP000075900 UP000075920 UP000075882 UP000007819 UP000094527 UP000075884
UP000007266 UP000192223 UP000019118 UP000030742 UP000242457 UP000005203 UP000092553 UP000053097 UP000279307 UP000001070 UP000036403 UP000076502 UP000268350 UP000007798 UP000009192 UP000001819 UP000008237 UP000192221 UP000095300 UP000007801 UP000008792 UP000037069 UP000000311 UP000095301 UP000001292 UP000078542 UP000078492 UP000007755 UP000078540 UP000078541 UP000005205 UP000075809 UP000000803 UP000002282 UP000092443 UP000092460 UP000008711 UP000092445 UP000078200 UP000235965 UP000008820 UP000053105 UP000069940 UP000075880 UP000092444 UP000075903 UP000075840 UP000076407 UP000075885 UP000075902 UP000075886 UP000069272 UP000007062 UP000283509 UP000000673 UP000075883 UP000076408 UP000075900 UP000075920 UP000075882 UP000007819 UP000094527 UP000075884
Interpro
ProteinModelPortal
H9JE78
A0A3S2LI51
A0A2H1W3L6
A0A2A4K0M3
A0A194PIV0
A0A0N1PJR3
+ More
A0A1E1WKG0 A0A212FKU9 A0A1Y1LXZ9 D6X4J2 A0A1W4XGA9 N6SWJ6 A0A1L8DWE5 A0A1L8DVV8 W8BVN0 A0A2A3EB91 A0A088A2H6 U5EXH0 A0A0M5J2Y4 A0A0A1WQU2 A0A026WEC6 A0A034VAK3 A0A310SW74 B4JMT2 A0A0J7L0I5 A0A0K8VW77 A0A154NY20 A0A336MTI7 A0A3B0K3M6 B4NEG2 B4L566 Q29GY7 E2BA54 A0A1W4UW73 A0A1I8NYC1 B3MQ12 E9IFD1 B4M9W1 A0A0L0BU93 E2A3P5 A0A0T6B4N0 A0A1I8M1E4 B4IHI7 A0A151IJS6 A0A151JB39 F4WWC5 A0A195B073 A0A195FCM0 A0A158NKJ6 A0A151WRP9 O76928 B4Q159 A0A1A9XRV6 A0A1B0AP73 B3NXY4 A0A1A9ZE61 A0A1A9UUP0 A0A336LZ39 A0A1S4FTJ5 A0A2J7R1C3 A0A1B6K793 Q0IEC0 A0A0N1IT56 A0A1B6GN75 A0A182GZT4 A0A182IMQ8 A0A1B0FQM7 A0A182V0N7 A0A0A9WH95 A0A1B6KHT4 A0A224XD99 A0A023F5N5 A0A182HL04 A0A069DU23 A0A182XA25 A0A182PH03 A0A182UJQ2 A0A1B6CS50 A0A182QEN2 A0A336MWC8 A0A2M3Z9D8 A0A182FRS3 A0A2M4BPW4 A0A0V0G7J8 A0A2M4ANP9 Q7PXM8 A0A423SW68 A0A2M3Z934 W5JHX4 A0A182MJC8 A0A182YB98 A0A0P4VTB8 A0A182R514 A0A182VWD3 A0A182L9C4 J9JYV0 A0A1D2N4A9 A0A182N547 A0A336LZ45 A0A2H8U036
A0A1E1WKG0 A0A212FKU9 A0A1Y1LXZ9 D6X4J2 A0A1W4XGA9 N6SWJ6 A0A1L8DWE5 A0A1L8DVV8 W8BVN0 A0A2A3EB91 A0A088A2H6 U5EXH0 A0A0M5J2Y4 A0A0A1WQU2 A0A026WEC6 A0A034VAK3 A0A310SW74 B4JMT2 A0A0J7L0I5 A0A0K8VW77 A0A154NY20 A0A336MTI7 A0A3B0K3M6 B4NEG2 B4L566 Q29GY7 E2BA54 A0A1W4UW73 A0A1I8NYC1 B3MQ12 E9IFD1 B4M9W1 A0A0L0BU93 E2A3P5 A0A0T6B4N0 A0A1I8M1E4 B4IHI7 A0A151IJS6 A0A151JB39 F4WWC5 A0A195B073 A0A195FCM0 A0A158NKJ6 A0A151WRP9 O76928 B4Q159 A0A1A9XRV6 A0A1B0AP73 B3NXY4 A0A1A9ZE61 A0A1A9UUP0 A0A336LZ39 A0A1S4FTJ5 A0A2J7R1C3 A0A1B6K793 Q0IEC0 A0A0N1IT56 A0A1B6GN75 A0A182GZT4 A0A182IMQ8 A0A1B0FQM7 A0A182V0N7 A0A0A9WH95 A0A1B6KHT4 A0A224XD99 A0A023F5N5 A0A182HL04 A0A069DU23 A0A182XA25 A0A182PH03 A0A182UJQ2 A0A1B6CS50 A0A182QEN2 A0A336MWC8 A0A2M3Z9D8 A0A182FRS3 A0A2M4BPW4 A0A0V0G7J8 A0A2M4ANP9 Q7PXM8 A0A423SW68 A0A2M3Z934 W5JHX4 A0A182MJC8 A0A182YB98 A0A0P4VTB8 A0A182R514 A0A182VWD3 A0A182L9C4 J9JYV0 A0A1D2N4A9 A0A182N547 A0A336LZ45 A0A2H8U036
PDB
4IKN
E-value=8.96025e-106,
Score=980
Ontologies
GO
Topology
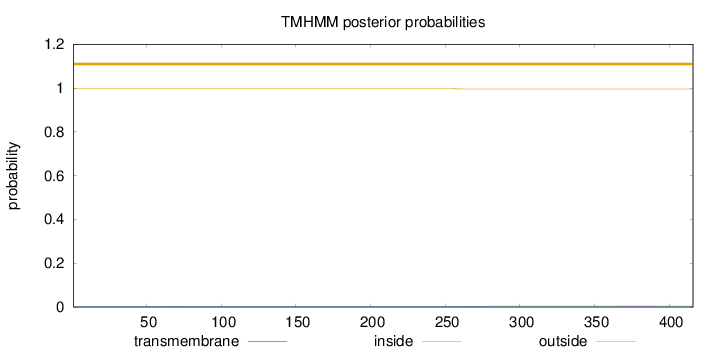
Length:
416
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0501400000000001
Exp number, first 60 AAs:
0.00032
Total prob of N-in:
0.00219
outside
1 - 416
Population Genetic Test Statistics
Pi
227.917363
Theta
188.510392
Tajima's D
0.777888
CLR
0
CSRT
0.591720413979301
Interpretation
Uncertain
