Gene
KWMTBOMO08889 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007829
Annotation
PREDICTED:_uncharacterized_protein_LOC106138721_[Amyelois_transitella]
Location in the cell
Extracellular Reliability : 2.672
Sequence
CDS
ATGAAGAGCAAGGCGCTTAGCTTGGCGCCTTTAGCACTGTTGCTACTAATACAAGCTTCATCTGTACTTAGTCGGATAGCGTTCGAGAAACTGACGGATGTGGACTTCAGTGGTACTGCGTATTACACCGTGCGGAACCTGTCCTTATACGAGTGCCAGGGCTGGTGCCGAGAGGAACCTGATTGTCAGGCGGCTTCCTTCAGTTTCGTCCTCAATCCTCTAACGCCCGTGCAGGAAACCAGATGTCTGCTACAAAACGATACACAGGCCAATAACCCCATGGCTACACCGCAGCGTTCCGTGAACACTTACTACATGGTTAAGCTGCAAGTACGCTCGGAGAGTGTATGCTTGCGGCCATGGGCATTCGAAAGAGTACCAGGGAAATCACTCCGTGGTCTTGACAACACGATCATCTACACTTCAACTAAGGAAGCGTGTTTGGCTGCGTGTCTCAATGAGAAAAAGTTACCGTGCAGATCCGCAGAATATGAGTACGGAAGCATGAGATGTGCCCTGAGTGATTCCGACAGACGCACCGCTCAACATTTTGTTCAACTAACCGATACCCCTGGCACCGATTACTTTGAGAACTTGTGCCTGAAGGCTGGACAGGCCTGTAAGGGCCAACGCGTCTTCACCGCGCCGCGCGTAGGAGTCGCCGAAGATAAGGTTGCTCAGTATGCAGGACTGCATTACTACACCGACAAAGAGTTACAAGTGACGTCAGAGGCGGCTTGTCGGCTTGCATGCGAAATCGAATCTGAGTTCCTGTGCCGATCGTTCCTCTATCTCGGAGCTCCGCACTCCGCTGCCTACAACTGCAGACTGTACCACCTCGACCACCACACGCTGCCCGACGGTCCTTCCGCCTATCTGAACGCCGAAAGACCTTTGATCGACGACGGCGAACCGATCGGAAAGTACTCCGAGAACTTCTGCGAGAAACCGGCGAGCAATGCGAACCCGCCCGGAGAGTTACCTGTGACGATCGAACACCAGCAAGACACGAACATGTCTAGCAACCTCACCAGGAACGATGCTAATTGCGACAAAACGGGAACATGTTACGACGTGTCGGTGCTTTGCAAGGATACAAGGATCGCAGTGCAGGTCCGCACGAACAAGCCCTTCAACGGTCGGATCTACGCTTTGGGTCGCTCGGAAACATGCAACATTGATGTAGTCAATAGCGATCTTTTCCGCTTGGACCTCACCATGGCCGGACAGGACTGCAATACTCAGAGCGTGACTGGGGTATATTCAAACACCGTCGTACTGCAACACCACAGCGTGGTCATGACGAAAGCCGACAAGATCTACAAAGTCAAGTGCACGTACGACATGAGTTCGAAGAATATCACATTCGGAATGGTGCCGATCAGAGATCCAGAGATGATTTCGATCACGGCTTCTCCGGAAGCACCTCCACCGCGCATTCGCATTTTGGACACTCGTCAACGCGAGGTCGATTCTGTTCGCATCGGAGACAGCCTCGCCTTCCGTATCGAGATCCCGGAAGACACCCCTTACGGTATCTTCGCTCGCAGCTGTGTCGCTATGGCTAAAGACTCCAAGACCACATTCCAGATTATTGACGACGAAGGATGTCCGGTCGATCCCGCTATATTCCCAGGTTTTACTCCTGATGGCAACGCGCTGCAGTCGATTTACGAAGCCTTTAGATTTACCGAGTCTTACGGTGTAATCTTCCAATGTAATGTCAAGTATTGTCTTGGACCTTGTGAGCCGGCGGTGTGCGAATGGGGCAGAGATTCTCTCGAGTCTTGGGGAAGAAAGAAGCGTTCGCTTCCTCGCAACGACACCGAAGGCCCCGCTCAGGACGAAGAAATGAATATCTCCCAAGAAATTTTAGTCTTGGACTTTGGTGATGAGCATCAGAGCACTGATTTTCTACGTTCTGACAAGCCCGGCGGAGTTGCTTCCGAGGCCAACTTTGGAGAAAAAACGGTCACGATCGTGGAGCCCTGTCCCAGTAAAGCTTCAGTGCTCCTGCTCGGAGTCGCGTGCGCGTTGCTCGTCCTTCTCTACATAGCGACGATCTTCTGCTACTACATGCGCAAGTGGCTGACTCCGCCGAAGCACATGTCGTAA
Protein
MKSKALSLAPLALLLLIQASSVLSRIAFEKLTDVDFSGTAYYTVRNLSLYECQGWCREEPDCQAASFSFVLNPLTPVQETRCLLQNDTQANNPMATPQRSVNTYYMVKLQVRSESVCLRPWAFERVPGKSLRGLDNTIIYTSTKEACLAACLNEKKLPCRSAEYEYGSMRCALSDSDRRTAQHFVQLTDTPGTDYFENLCLKAGQACKGQRVFTAPRVGVAEDKVAQYAGLHYYTDKELQVTSEAACRLACEIESEFLCRSFLYLGAPHSAAYNCRLYHLDHHTLPDGPSAYLNAERPLIDDGEPIGKYSENFCEKPASNANPPGELPVTIEHQQDTNMSSNLTRNDANCDKTGTCYDVSVLCKDTRIAVQVRTNKPFNGRIYALGRSETCNIDVVNSDLFRLDLTMAGQDCNTQSVTGVYSNTVVLQHHSVVMTKADKIYKVKCTYDMSSKNITFGMVPIRDPEMISITASPEAPPPRIRILDTRQREVDSVRIGDSLAFRIEIPEDTPYGIFARSCVAMAKDSKTTFQIIDDEGCPVDPAIFPGFTPDGNALQSIYEAFRFTESYGVIFQCNVKYCLGPCEPAVCEWGRDSLESWGRKKRSLPRNDTEGPAQDEEMNISQEILVLDFGDEHQSTDFLRSDKPGGVASEANFGEKTVTIVEPCPSKASVLLLGVACALLVLLYIATIFCYYMRKWLTPPKHMS
Summary
Uniprot
A0A2H1WDX9
A0A3S2NRB4
A0A194PQ35
A0A194RDJ8
A0A212FKV4
D6WCQ0
+ More
A0A0L7LA69 A0A139WM22 A0A1Y1KDA6 A0A1W4XI26 A0A067RG78 A0A151WFX6 E2BV16 A0A2R7WJA5 A0A2S2PWP9 A0A0L7R538 A0A154P8R1 A0A151INZ7 A0A195BU26 A0A088AC97 F4X815 A0A195FXK7 A0A158NUD3 A0A151J1A2 J9K0U0 A0A1S3D587 A0A2H8TNX4 E2AEN9 W5J6L5 A0A182P5T7 T1I1K7 A0A182V7G1 A0A182LFU3 A0A182WRJ9 Q7QD96 A0A182HU77 A0A182IT52 N6TNG4 A0A182RNR5 A0A182QEI7 A0A182TPR8 A0A1S4FG50 A0A182M333 A0A182Y4A5 A0A182W062 A0A0A9ZB53 A0A182KAY0 A0A182T015 B4NCY5 A0A182N642 A0A1B0CDT4 A0A1I8NPE1 A0A182FRE6 B4PWJ2 B4MEP6 A0A026VZ91 B4JMU5 A0A0A1X1C0 A0A0K8U7H8 A0A0R1E8R0 A0A034V269 A0A0R1E956 B3P944 A0A1W4V9D3 B4L2D3 A0A0L0C513 Q8MS37 A0A0R1EAK2 A0A0C9Q6E7 A0A3L8DP99 A0A0R1E9F3 A0A3B0KS22 B4H315 B5DKG8 B4R722 A0A1I8M8N5 B4IM31 B3MYN7 A0A1J1IGE1 E0VC84 A0A336MT58 A0A1B0FMH8 A0A1A9Z0F1 A0A1A9UT06 A0A1B0B615 A0A1A9XLY9 A0A1A9X5N1 A0A0A1XIZ1 K7IRC8 A0A232F4N8 A0A310SLW1 A0A0M4EXF3 A0A0K8W7I2 A0A336MYJ1 A0A1B0D9P3 A0A0K8URT7 A0A2A4IXP5 A0A2A3EFQ3 A0A2P8XY43 E9J910
A0A0L7LA69 A0A139WM22 A0A1Y1KDA6 A0A1W4XI26 A0A067RG78 A0A151WFX6 E2BV16 A0A2R7WJA5 A0A2S2PWP9 A0A0L7R538 A0A154P8R1 A0A151INZ7 A0A195BU26 A0A088AC97 F4X815 A0A195FXK7 A0A158NUD3 A0A151J1A2 J9K0U0 A0A1S3D587 A0A2H8TNX4 E2AEN9 W5J6L5 A0A182P5T7 T1I1K7 A0A182V7G1 A0A182LFU3 A0A182WRJ9 Q7QD96 A0A182HU77 A0A182IT52 N6TNG4 A0A182RNR5 A0A182QEI7 A0A182TPR8 A0A1S4FG50 A0A182M333 A0A182Y4A5 A0A182W062 A0A0A9ZB53 A0A182KAY0 A0A182T015 B4NCY5 A0A182N642 A0A1B0CDT4 A0A1I8NPE1 A0A182FRE6 B4PWJ2 B4MEP6 A0A026VZ91 B4JMU5 A0A0A1X1C0 A0A0K8U7H8 A0A0R1E8R0 A0A034V269 A0A0R1E956 B3P944 A0A1W4V9D3 B4L2D3 A0A0L0C513 Q8MS37 A0A0R1EAK2 A0A0C9Q6E7 A0A3L8DP99 A0A0R1E9F3 A0A3B0KS22 B4H315 B5DKG8 B4R722 A0A1I8M8N5 B4IM31 B3MYN7 A0A1J1IGE1 E0VC84 A0A336MT58 A0A1B0FMH8 A0A1A9Z0F1 A0A1A9UT06 A0A1B0B615 A0A1A9XLY9 A0A1A9X5N1 A0A0A1XIZ1 K7IRC8 A0A232F4N8 A0A310SLW1 A0A0M4EXF3 A0A0K8W7I2 A0A336MYJ1 A0A1B0D9P3 A0A0K8URT7 A0A2A4IXP5 A0A2A3EFQ3 A0A2P8XY43 E9J910
Pubmed
26354079
22118469
18362917
19820115
26227816
28004739
+ More
24845553 20798317 21719571 21347285 20920257 23761445 20966253 12364791 14747013 17210077 23537049 25244985 25401762 26823975 17994087 17550304 24508170 25830018 25348373 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 30249741 15632085 23185243 25315136 20566863 20075255 28648823 29403074 21282665
24845553 20798317 21719571 21347285 20920257 23761445 20966253 12364791 14747013 17210077 23537049 25244985 25401762 26823975 17994087 17550304 24508170 25830018 25348373 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 30249741 15632085 23185243 25315136 20566863 20075255 28648823 29403074 21282665
EMBL
ODYU01008024
SOQ51285.1
RSAL01000005
RVE54345.1
KQ459602
KPI93245.1
+ More
KQ460367 KPJ15534.1 AGBW02007963 OWR54364.1 KQ971317 EEZ97801.2 JTDY01002003 KOB72357.1 KYB28982.1 GEZM01090020 JAV57386.1 KK852703 KDR18083.1 KQ983203 KYQ46729.1 GL450786 EFN80467.1 KK854832 PTY19100.1 GGMS01000581 MBY69784.1 KQ414654 KOC66000.1 KQ434845 KZC08231.1 KQ976899 KYN07194.1 KQ976417 KYM89919.1 GL888900 EGI57482.1 KQ981193 KYN45168.1 ADTU01026379 KQ980575 KYN15508.1 ABLF02034919 ABLF02034926 GFXV01003924 MBW15729.1 GL438870 EFN68114.1 ADMH02002131 ETN58495.1 ACPB03005172 AAAB01008859 EAA07921.4 APCN01000611 APCN01000612 APGK01017071 APGK01017072 APGK01017073 APGK01017074 APGK01017075 APGK01017076 APGK01017077 APGK01017078 APGK01017079 APGK01017080 KB739995 ENN82034.1 AXCN02000194 AXCM01006253 GBHO01003022 GBHO01003021 GDHC01019877 JAG40582.1 JAG40583.1 JAP98751.1 CH964239 EDW82694.1 AJWK01008212 AJWK01008213 CM000162 EDX00761.2 CH940664 EDW63021.2 KK107539 EZA49098.1 CH916371 EDV92038.1 GBXI01009153 GBXI01003927 JAD05139.1 JAD10365.1 GDHF01030039 GDHF01029108 GDHF01028437 GDHF01016239 GDHF01003790 JAI22275.1 JAI23206.1 JAI23877.1 JAI36075.1 JAI48524.1 KRK05776.1 GAKP01021583 GAKP01021581 JAC37371.1 KRK05773.1 CH954183 EDV45340.1 CH933810 EDW07794.1 JRES01000981 KNC26524.1 AY119115 AE014298 AAM50975.1 AAZ52488.1 ABC67158.1 AHN59198.1 AHN59199.1 AHN59200.1 AHN59201.1 KRK05774.1 GBYB01008933 GBYB01009683 JAG78700.1 JAG79450.1 QOIP01000006 RLU22116.1 KRK05775.1 KRK05777.1 OUUW01000011 SPP86728.1 CH479205 EDW30708.1 CH379063 EDY72017.1 KRT05841.1 KRT05842.1 KRT05843.1 CM000366 EDX16733.1 CH480923 EDW43658.1 CH902632 EDV32731.2 CVRI01000050 CRK99275.1 DS235048 EEB10990.1 UFQT01002592 SSX33734.1 CCAG010017550 JXJN01008943 GBXI01003311 JAD10981.1 NNAY01001023 OXU25449.1 KQ759886 OAD62134.1 CP012528 ALC48405.1 GDHF01016513 GDHF01015288 GDHF01005277 GDHF01001848 GDHF01001819 JAI35801.1 JAI37026.1 JAI47037.1 JAI50466.1 JAI50495.1 UFQT01003183 SSX34711.1 AJVK01028362 AJVK01028363 GDHF01022900 GDHF01012997 JAI29414.1 JAI39317.1 NWSH01005112 PCG64429.1 KZ288256 PBC30585.1 PYGN01001182 PSN36834.1 GL769168 EFZ10688.1
KQ460367 KPJ15534.1 AGBW02007963 OWR54364.1 KQ971317 EEZ97801.2 JTDY01002003 KOB72357.1 KYB28982.1 GEZM01090020 JAV57386.1 KK852703 KDR18083.1 KQ983203 KYQ46729.1 GL450786 EFN80467.1 KK854832 PTY19100.1 GGMS01000581 MBY69784.1 KQ414654 KOC66000.1 KQ434845 KZC08231.1 KQ976899 KYN07194.1 KQ976417 KYM89919.1 GL888900 EGI57482.1 KQ981193 KYN45168.1 ADTU01026379 KQ980575 KYN15508.1 ABLF02034919 ABLF02034926 GFXV01003924 MBW15729.1 GL438870 EFN68114.1 ADMH02002131 ETN58495.1 ACPB03005172 AAAB01008859 EAA07921.4 APCN01000611 APCN01000612 APGK01017071 APGK01017072 APGK01017073 APGK01017074 APGK01017075 APGK01017076 APGK01017077 APGK01017078 APGK01017079 APGK01017080 KB739995 ENN82034.1 AXCN02000194 AXCM01006253 GBHO01003022 GBHO01003021 GDHC01019877 JAG40582.1 JAG40583.1 JAP98751.1 CH964239 EDW82694.1 AJWK01008212 AJWK01008213 CM000162 EDX00761.2 CH940664 EDW63021.2 KK107539 EZA49098.1 CH916371 EDV92038.1 GBXI01009153 GBXI01003927 JAD05139.1 JAD10365.1 GDHF01030039 GDHF01029108 GDHF01028437 GDHF01016239 GDHF01003790 JAI22275.1 JAI23206.1 JAI23877.1 JAI36075.1 JAI48524.1 KRK05776.1 GAKP01021583 GAKP01021581 JAC37371.1 KRK05773.1 CH954183 EDV45340.1 CH933810 EDW07794.1 JRES01000981 KNC26524.1 AY119115 AE014298 AAM50975.1 AAZ52488.1 ABC67158.1 AHN59198.1 AHN59199.1 AHN59200.1 AHN59201.1 KRK05774.1 GBYB01008933 GBYB01009683 JAG78700.1 JAG79450.1 QOIP01000006 RLU22116.1 KRK05775.1 KRK05777.1 OUUW01000011 SPP86728.1 CH479205 EDW30708.1 CH379063 EDY72017.1 KRT05841.1 KRT05842.1 KRT05843.1 CM000366 EDX16733.1 CH480923 EDW43658.1 CH902632 EDV32731.2 CVRI01000050 CRK99275.1 DS235048 EEB10990.1 UFQT01002592 SSX33734.1 CCAG010017550 JXJN01008943 GBXI01003311 JAD10981.1 NNAY01001023 OXU25449.1 KQ759886 OAD62134.1 CP012528 ALC48405.1 GDHF01016513 GDHF01015288 GDHF01005277 GDHF01001848 GDHF01001819 JAI35801.1 JAI37026.1 JAI47037.1 JAI50466.1 JAI50495.1 UFQT01003183 SSX34711.1 AJVK01028362 AJVK01028363 GDHF01022900 GDHF01012997 JAI29414.1 JAI39317.1 NWSH01005112 PCG64429.1 KZ288256 PBC30585.1 PYGN01001182 PSN36834.1 GL769168 EFZ10688.1
Proteomes
UP000283053
UP000053268
UP000053240
UP000007151
UP000007266
UP000037510
+ More
UP000192223 UP000027135 UP000075809 UP000008237 UP000053825 UP000076502 UP000078542 UP000078540 UP000005203 UP000007755 UP000078541 UP000005205 UP000078492 UP000007819 UP000079169 UP000000311 UP000000673 UP000075885 UP000015103 UP000075903 UP000075882 UP000076407 UP000007062 UP000075840 UP000075880 UP000019118 UP000075900 UP000075886 UP000075902 UP000075883 UP000076408 UP000075920 UP000075881 UP000075901 UP000007798 UP000075884 UP000092461 UP000095300 UP000069272 UP000002282 UP000008792 UP000053097 UP000001070 UP000008711 UP000192221 UP000009192 UP000037069 UP000000803 UP000279307 UP000268350 UP000008744 UP000001819 UP000000304 UP000095301 UP000001292 UP000007801 UP000183832 UP000009046 UP000092444 UP000092445 UP000078200 UP000092460 UP000092443 UP000091820 UP000002358 UP000215335 UP000092553 UP000092462 UP000218220 UP000242457 UP000245037
UP000192223 UP000027135 UP000075809 UP000008237 UP000053825 UP000076502 UP000078542 UP000078540 UP000005203 UP000007755 UP000078541 UP000005205 UP000078492 UP000007819 UP000079169 UP000000311 UP000000673 UP000075885 UP000015103 UP000075903 UP000075882 UP000076407 UP000007062 UP000075840 UP000075880 UP000019118 UP000075900 UP000075886 UP000075902 UP000075883 UP000076408 UP000075920 UP000075881 UP000075901 UP000007798 UP000075884 UP000092461 UP000095300 UP000069272 UP000002282 UP000008792 UP000053097 UP000001070 UP000008711 UP000192221 UP000009192 UP000037069 UP000000803 UP000279307 UP000268350 UP000008744 UP000001819 UP000000304 UP000095301 UP000001292 UP000007801 UP000183832 UP000009046 UP000092444 UP000092445 UP000078200 UP000092460 UP000092443 UP000091820 UP000002358 UP000215335 UP000092553 UP000092462 UP000218220 UP000242457 UP000245037
PRIDE
ProteinModelPortal
A0A2H1WDX9
A0A3S2NRB4
A0A194PQ35
A0A194RDJ8
A0A212FKV4
D6WCQ0
+ More
A0A0L7LA69 A0A139WM22 A0A1Y1KDA6 A0A1W4XI26 A0A067RG78 A0A151WFX6 E2BV16 A0A2R7WJA5 A0A2S2PWP9 A0A0L7R538 A0A154P8R1 A0A151INZ7 A0A195BU26 A0A088AC97 F4X815 A0A195FXK7 A0A158NUD3 A0A151J1A2 J9K0U0 A0A1S3D587 A0A2H8TNX4 E2AEN9 W5J6L5 A0A182P5T7 T1I1K7 A0A182V7G1 A0A182LFU3 A0A182WRJ9 Q7QD96 A0A182HU77 A0A182IT52 N6TNG4 A0A182RNR5 A0A182QEI7 A0A182TPR8 A0A1S4FG50 A0A182M333 A0A182Y4A5 A0A182W062 A0A0A9ZB53 A0A182KAY0 A0A182T015 B4NCY5 A0A182N642 A0A1B0CDT4 A0A1I8NPE1 A0A182FRE6 B4PWJ2 B4MEP6 A0A026VZ91 B4JMU5 A0A0A1X1C0 A0A0K8U7H8 A0A0R1E8R0 A0A034V269 A0A0R1E956 B3P944 A0A1W4V9D3 B4L2D3 A0A0L0C513 Q8MS37 A0A0R1EAK2 A0A0C9Q6E7 A0A3L8DP99 A0A0R1E9F3 A0A3B0KS22 B4H315 B5DKG8 B4R722 A0A1I8M8N5 B4IM31 B3MYN7 A0A1J1IGE1 E0VC84 A0A336MT58 A0A1B0FMH8 A0A1A9Z0F1 A0A1A9UT06 A0A1B0B615 A0A1A9XLY9 A0A1A9X5N1 A0A0A1XIZ1 K7IRC8 A0A232F4N8 A0A310SLW1 A0A0M4EXF3 A0A0K8W7I2 A0A336MYJ1 A0A1B0D9P3 A0A0K8URT7 A0A2A4IXP5 A0A2A3EFQ3 A0A2P8XY43 E9J910
A0A0L7LA69 A0A139WM22 A0A1Y1KDA6 A0A1W4XI26 A0A067RG78 A0A151WFX6 E2BV16 A0A2R7WJA5 A0A2S2PWP9 A0A0L7R538 A0A154P8R1 A0A151INZ7 A0A195BU26 A0A088AC97 F4X815 A0A195FXK7 A0A158NUD3 A0A151J1A2 J9K0U0 A0A1S3D587 A0A2H8TNX4 E2AEN9 W5J6L5 A0A182P5T7 T1I1K7 A0A182V7G1 A0A182LFU3 A0A182WRJ9 Q7QD96 A0A182HU77 A0A182IT52 N6TNG4 A0A182RNR5 A0A182QEI7 A0A182TPR8 A0A1S4FG50 A0A182M333 A0A182Y4A5 A0A182W062 A0A0A9ZB53 A0A182KAY0 A0A182T015 B4NCY5 A0A182N642 A0A1B0CDT4 A0A1I8NPE1 A0A182FRE6 B4PWJ2 B4MEP6 A0A026VZ91 B4JMU5 A0A0A1X1C0 A0A0K8U7H8 A0A0R1E8R0 A0A034V269 A0A0R1E956 B3P944 A0A1W4V9D3 B4L2D3 A0A0L0C513 Q8MS37 A0A0R1EAK2 A0A0C9Q6E7 A0A3L8DP99 A0A0R1E9F3 A0A3B0KS22 B4H315 B5DKG8 B4R722 A0A1I8M8N5 B4IM31 B3MYN7 A0A1J1IGE1 E0VC84 A0A336MT58 A0A1B0FMH8 A0A1A9Z0F1 A0A1A9UT06 A0A1B0B615 A0A1A9XLY9 A0A1A9X5N1 A0A0A1XIZ1 K7IRC8 A0A232F4N8 A0A310SLW1 A0A0M4EXF3 A0A0K8W7I2 A0A336MYJ1 A0A1B0D9P3 A0A0K8URT7 A0A2A4IXP5 A0A2A3EFQ3 A0A2P8XY43 E9J910
Ontologies
Topology
SignalP
Position: 1 - 24,
Likelihood: 0.995224
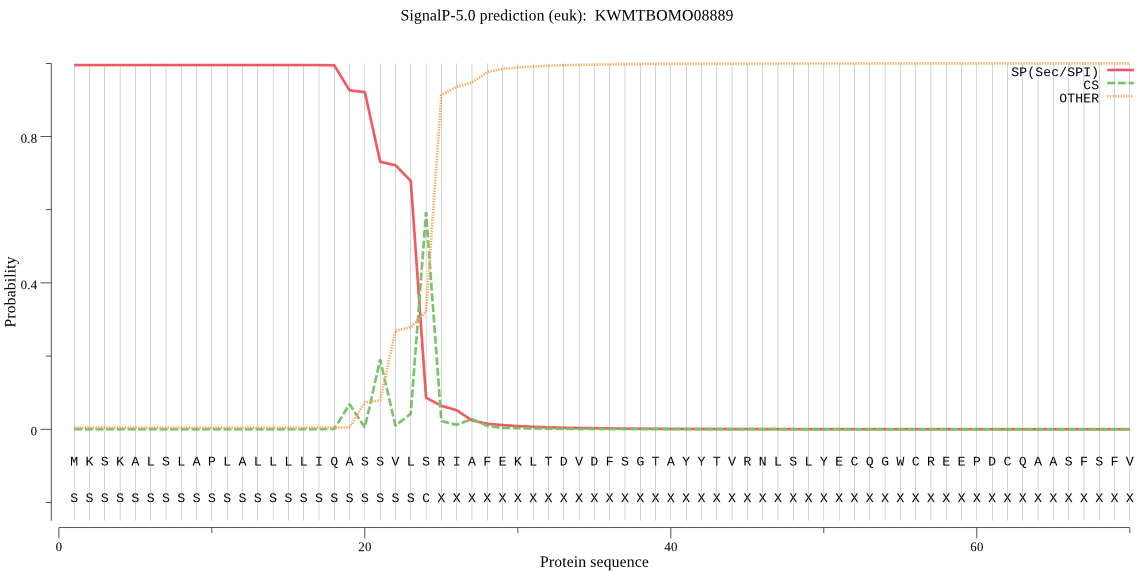
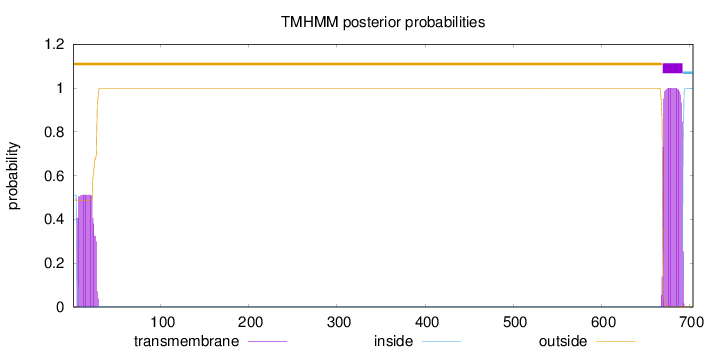
Length:
704
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
33.67454
Exp number, first 60 AAs:
10.83127
Total prob of N-in:
0.51159
POSSIBLE N-term signal
sequence
outside
1 - 669
TMhelix
670 - 692
inside
693 - 704
Population Genetic Test Statistics
Pi
266.610018
Theta
187.63421
Tajima's D
1.18444
CLR
0.313849
CSRT
0.704714764261787
Interpretation
Uncertain
