Gene
KWMTBOMO08888
Annotation
putative_lipoate-protein_ligase_b_[Danaus_plexippus]
Full name
Putative lipoyltransferase 2, mitochondrial
Alternative Name
Lipoate-protein ligase B
Lipoyl/octanoyl transferase
Octanoyl-[acyl-carrier-protein]-protein N-octanoyltransferase
Lipoyl/octanoyl transferase
Octanoyl-[acyl-carrier-protein]-protein N-octanoyltransferase
Location in the cell
Cytoplasmic Reliability : 1.84 Mitochondrial Reliability : 1.86
Sequence
CDS
ATGGTTAAAATTTGGAGGCTGGGTTTAATGAGCTATGATACTGCTCTTAATATCCAATTAGCTATAGCACGAAAACATCTCGATTCAATTAAGAATGGCCTGGATTTTAATCATGATACCCTTTTACTTGTAGAACATAAACCTGTTTATACAGTAGGTATAAGAGACAAGACACCAAACAATGAAATTCTCAGGCTAAAAAGTCTGGGAGCAGATTTTAAGAAAACAAACAGAGGCGGTCTTGTAACTTTTCATGGGCCTGGTCAATTAGTTGCATACCCAATTATAAATTTGAAGTACTATAAACCAAGTGTGAAGTGGTATGTAAATAGTTTGGAAGAAATTGGCATAAGATTGTGTGGTGACTTAGGAATTCAAGCCAGCAGATCACCTCACACAGGAGTATGGGTGGCAGATAACAAAATAGCCGCAATAGGTGTTCACACCTCGAGATATGTCACAACCCATGGTATTGCAATAAATTGTGATATTGACCTTTCTTGGTTCAAACACATAGACCCCTGTGGTATAGTTGACAAAGGAGTTACATCATTGACCAAAGAAACAGGAGATTTGTGCACAATAGACAAAATAACACCATTGTTTTTGAGTAAATTTCAAGATGTGTTTGAATGTGAGATAGAAGACCTGGACTTGAAGTCCCAAAAAGATATTCTGAGTTATGTATATAATAAATTAATGAAGGAATTGATGGTACAAAATACAGCATAA
Protein
MVKIWRLGLMSYDTALNIQLAIARKHLDSIKNGLDFNHDTLLLVEHKPVYTVGIRDKTPNNEILRLKSLGADFKKTNRGGLVTFHGPGQLVAYPIINLKYYKPSVKWYVNSLEEIGIRLCGDLGIQASRSPHTGVWVADNKIAAIGVHTSRYVTTHGIAINCDIDLSWFKHIDPCGIVDKGVTSLTKETGDLCTIDKITPLFLSKFQDVFECEIEDLDLKSQKDILSYVYNKLMKELMVQNTA
Summary
Description
Catalyzes the transfer of endogenously produced octanoic acid from octanoyl-acyl-carrier-protein onto the lipoyl domains of lipoate-dependent enzymes. Lipoyl-ACP can also act as a substrate although octanoyl-ACP is likely to be the physiological substrate.
Catalyzes the transfer of endogenously produced octanoic acid from octanoyl-acyl-carrier-protein onto the lipoyl domains of lipoate-dependent enzymes, which catalyze essential redox reactions (By similarity). Lipoyl-ACP can also act as a substrate although octanoyl-ACP is likely to be the physiological substrate (By similarity).
Catalyzes the transfer of endogenously produced octanoic acid from octanoyl-acyl-carrier-protein onto the lipoyl domains of lipoate-dependent enzymes, which catalyze essential redox reactions (By similarity). Lipoyl-ACP can also act as a substrate although octanoyl-ACP is likely to be the physiological substrate (By similarity).
Catalytic Activity
L-lysyl-[protein] + octanoyl-[ACP] = H(+) + holo-[ACP] + N(6)-octanoyl-L-lysyl-[protein]
Miscellaneous
In the reaction, the free carboxyl group of octanoic acid is attached via an amide linkage to the epsilon-amino group of a specific lysine residue of lipoyl domains of lipoate-dependent enzymes.
Similarity
Belongs to the LipB family.
Keywords
Acyltransferase
Complete proteome
Ligase
Mitochondrion
Reference proteome
Transferase
Transit peptide
Feature
chain Putative lipoyltransferase 2, mitochondrial
Uniprot
A0A212FKU4
A0A2A4IZ19
A0A2H1WEA6
A0A194RE29
A0A194PKI5
A0A437BVD1
+ More
A0A0L7LAW1 A0A2J7QYL2 A0A0L7QY58 E2A2V5 A0A2P8XZI4 A0A3P8YKA1 A0A2T7NCM6 A0A088ALW7 A0A151I3F1 A0A2A3E6B0 A0A154NWE4 C1BYA8 A0A158NXY6 U4U015 H3A1P6 A0A1W4YTY4 Q0IEK7 A0A0J7L363 N6T2D6 A0A060WM11 A0A3B1KFN2 A0A0P7UV33 E3TGJ5 F4WMW4 A0A067RKW3 A0A3B3CEV7 A0A3B1JQ22 A0A1S4FN42 A0A151WIG7 A0A3B4DS71 A0A151IVW7 A0A2D0SSE5 A0A0P6FSE3 A0A3N0XI49 E2BQH3 A0A0P5KBG0 A0A1B6LDI8 A0A0P6BGB1 A0A1A9UZU6 A0A0M9A1L5 E9IID1 A0A2R7WBM1 B0WH35 A0A1S3NPT8 D6WCX6 K7IYB8 U5EXR5 W4XML7 A0A1B0AL12 A0A1A9XN22 A0A1B0FR83 A0A2C9JF27 A0A1B6HLI8 A0A3Q1ETW8 A0A437CTR0 A0A0L8FR25 A0A1A8P1U8 Q4SB31 A0A3Q1D0S5 A0A3P8TSF6 A0A182LXS1 A0A1A9ZZQ8 A0A026VU07 A0A1A8J639 A0A1A8C774 A0A1A7ZW42 A0A1B0CL88 A0A131XKJ9 A0A3B5B6N9 E0VS59 A0A1A8H0Q2 A0A1L8HBD3 A0A3Q4N1Z4 I3KKT0 A0A3M6UTA1 A0A0F8BU33 Q7PS31 A0A182XEE9 A0A182IAB4 Q29R99 R7V6C9 A0A1Q3EXD3 A0A1A8L678 A0A2M4CS08 A0A3B3QZL2 A0A3Q1J525 A0A1S3DW12 A0A1L8E0L8 F1QAW5 A0A131YV83 A0A182S5L7 A0A3Q2R0K8 A0A3P9MXY6 A0A3B4ZA36
A0A0L7LAW1 A0A2J7QYL2 A0A0L7QY58 E2A2V5 A0A2P8XZI4 A0A3P8YKA1 A0A2T7NCM6 A0A088ALW7 A0A151I3F1 A0A2A3E6B0 A0A154NWE4 C1BYA8 A0A158NXY6 U4U015 H3A1P6 A0A1W4YTY4 Q0IEK7 A0A0J7L363 N6T2D6 A0A060WM11 A0A3B1KFN2 A0A0P7UV33 E3TGJ5 F4WMW4 A0A067RKW3 A0A3B3CEV7 A0A3B1JQ22 A0A1S4FN42 A0A151WIG7 A0A3B4DS71 A0A151IVW7 A0A2D0SSE5 A0A0P6FSE3 A0A3N0XI49 E2BQH3 A0A0P5KBG0 A0A1B6LDI8 A0A0P6BGB1 A0A1A9UZU6 A0A0M9A1L5 E9IID1 A0A2R7WBM1 B0WH35 A0A1S3NPT8 D6WCX6 K7IYB8 U5EXR5 W4XML7 A0A1B0AL12 A0A1A9XN22 A0A1B0FR83 A0A2C9JF27 A0A1B6HLI8 A0A3Q1ETW8 A0A437CTR0 A0A0L8FR25 A0A1A8P1U8 Q4SB31 A0A3Q1D0S5 A0A3P8TSF6 A0A182LXS1 A0A1A9ZZQ8 A0A026VU07 A0A1A8J639 A0A1A8C774 A0A1A7ZW42 A0A1B0CL88 A0A131XKJ9 A0A3B5B6N9 E0VS59 A0A1A8H0Q2 A0A1L8HBD3 A0A3Q4N1Z4 I3KKT0 A0A3M6UTA1 A0A0F8BU33 Q7PS31 A0A182XEE9 A0A182IAB4 Q29R99 R7V6C9 A0A1Q3EXD3 A0A1A8L678 A0A2M4CS08 A0A3B3QZL2 A0A3Q1J525 A0A1S3DW12 A0A1L8E0L8 F1QAW5 A0A131YV83 A0A182S5L7 A0A3Q2R0K8 A0A3P9MXY6 A0A3B4ZA36
EC Number
2.3.1.181
Pubmed
22118469
26354079
26227816
20798317
29403074
25069045
+ More
20433749 21347285 23537049 9215903 17510324 24755649 25329095 20634964 21719571 24845553 29451363 21282665 18362917 19820115 20075255 15562597 15496914 24508170 30249741 28049606 20566863 27762356 25186727 30382153 25835551 12364791 14747013 17210077 23254933 29240929 23594743 26830274
20433749 21347285 23537049 9215903 17510324 24755649 25329095 20634964 21719571 24845553 29451363 21282665 18362917 19820115 20075255 15562597 15496914 24508170 30249741 28049606 20566863 27762356 25186727 30382153 25835551 12364791 14747013 17210077 23254933 29240929 23594743 26830274
EMBL
AGBW02007963
OWR54365.1
NWSH01005112
PCG64430.1
ODYU01008024
SOQ51286.1
+ More
KQ460367 KPJ15535.1 KQ459602 KPI93244.1 RSAL01000005 RVE54346.1 JTDY01002003 KOB72356.1 NEVH01009087 PNF33666.1 KQ414698 KOC63486.1 GL436217 EFN72229.1 PYGN01001121 PSN37421.1 PZQS01000014 PVD18929.1 KQ976482 KYM83711.1 KZ288353 PBC27303.1 KQ434773 KZC03999.1 BT079587 ACO14011.1 ADTU01003705 KB631924 ERL87224.1 AFYH01253371 AFYH01253372 AFYH01253373 AFYH01253374 AFYH01253375 CH477600 EAT38456.1 LBMM01001119 KMQ96889.1 APGK01055062 KB741259 ENN71698.1 FR904610 CDQ68087.1 JARO02002011 KPP73828.1 GU589481 ADO29431.1 GL888223 EGI64434.1 KK852617 KDR20162.1 KQ983089 KYQ47637.1 KQ980882 KYN11923.1 GDIQ01054879 JAN39858.1 RJVU01075616 ROI16089.1 GL449769 EFN82031.1 GDIQ01186524 JAK65201.1 GEBQ01018221 GEBQ01006588 GEBQ01004079 JAT21756.1 JAT33389.1 JAT35898.1 GDIP01017833 JAM85882.1 KQ435762 KOX75475.1 GL763459 EFZ19673.1 KK854583 PTY17092.1 DS231932 EDS27486.1 KQ971311 EEZ98826.2 GANO01002414 JAB57457.1 AAGJ04111440 JXJN01029586 CCAG010008016 GECU01032195 GECU01000519 JAS75511.1 JAT07188.1 CM012449 RVE65309.1 KQ427350 KOF67161.1 HAEH01004945 HAEI01006424 SBR75243.1 CAAE01014677 CAG02151.1 AXCM01000566 KK111586 QOIP01000007 EZA46339.1 RLU20153.1 HAED01018521 HAEE01005590 SBR04966.1 HADZ01010730 HAEA01008621 SBP74671.1 HADY01008617 HAEJ01012626 SBP47102.1 AJWK01017043 GEFH01002530 JAP66051.1 DS235745 EEB16215.1 HAEB01003734 HAEC01008739 SBQ76877.1 CM004469 OCT93420.1 AERX01047718 RCHS01000788 RMX56875.1 KQ042224 KKF17481.1 AAAB01008846 EAA06255.5 APCN01001139 BC114309 AMQN01004941 KB294746 ELU14124.1 GFDL01015187 JAV19858.1 HAEF01002765 HAEG01004064 SBR40147.1 GGFL01003929 MBW68107.1 GFDF01001847 JAV12237.1 CT027729 GEDV01006491 JAP82066.1
KQ460367 KPJ15535.1 KQ459602 KPI93244.1 RSAL01000005 RVE54346.1 JTDY01002003 KOB72356.1 NEVH01009087 PNF33666.1 KQ414698 KOC63486.1 GL436217 EFN72229.1 PYGN01001121 PSN37421.1 PZQS01000014 PVD18929.1 KQ976482 KYM83711.1 KZ288353 PBC27303.1 KQ434773 KZC03999.1 BT079587 ACO14011.1 ADTU01003705 KB631924 ERL87224.1 AFYH01253371 AFYH01253372 AFYH01253373 AFYH01253374 AFYH01253375 CH477600 EAT38456.1 LBMM01001119 KMQ96889.1 APGK01055062 KB741259 ENN71698.1 FR904610 CDQ68087.1 JARO02002011 KPP73828.1 GU589481 ADO29431.1 GL888223 EGI64434.1 KK852617 KDR20162.1 KQ983089 KYQ47637.1 KQ980882 KYN11923.1 GDIQ01054879 JAN39858.1 RJVU01075616 ROI16089.1 GL449769 EFN82031.1 GDIQ01186524 JAK65201.1 GEBQ01018221 GEBQ01006588 GEBQ01004079 JAT21756.1 JAT33389.1 JAT35898.1 GDIP01017833 JAM85882.1 KQ435762 KOX75475.1 GL763459 EFZ19673.1 KK854583 PTY17092.1 DS231932 EDS27486.1 KQ971311 EEZ98826.2 GANO01002414 JAB57457.1 AAGJ04111440 JXJN01029586 CCAG010008016 GECU01032195 GECU01000519 JAS75511.1 JAT07188.1 CM012449 RVE65309.1 KQ427350 KOF67161.1 HAEH01004945 HAEI01006424 SBR75243.1 CAAE01014677 CAG02151.1 AXCM01000566 KK111586 QOIP01000007 EZA46339.1 RLU20153.1 HAED01018521 HAEE01005590 SBR04966.1 HADZ01010730 HAEA01008621 SBP74671.1 HADY01008617 HAEJ01012626 SBP47102.1 AJWK01017043 GEFH01002530 JAP66051.1 DS235745 EEB16215.1 HAEB01003734 HAEC01008739 SBQ76877.1 CM004469 OCT93420.1 AERX01047718 RCHS01000788 RMX56875.1 KQ042224 KKF17481.1 AAAB01008846 EAA06255.5 APCN01001139 BC114309 AMQN01004941 KB294746 ELU14124.1 GFDL01015187 JAV19858.1 HAEF01002765 HAEG01004064 SBR40147.1 GGFL01003929 MBW68107.1 GFDF01001847 JAV12237.1 CT027729 GEDV01006491 JAP82066.1
Proteomes
UP000007151
UP000218220
UP000053240
UP000053268
UP000283053
UP000037510
+ More
UP000235965 UP000053825 UP000000311 UP000245037 UP000265140 UP000245119 UP000005203 UP000078540 UP000242457 UP000076502 UP000005205 UP000030742 UP000008672 UP000192224 UP000008820 UP000036403 UP000019118 UP000193380 UP000018467 UP000034805 UP000221080 UP000007755 UP000027135 UP000261560 UP000075809 UP000261440 UP000078492 UP000008237 UP000078200 UP000053105 UP000002320 UP000087266 UP000007266 UP000002358 UP000007110 UP000092460 UP000092443 UP000092444 UP000076420 UP000257200 UP000053454 UP000007303 UP000257160 UP000265080 UP000075883 UP000092445 UP000053097 UP000279307 UP000092461 UP000261400 UP000009046 UP000186698 UP000261580 UP000005207 UP000275408 UP000007062 UP000076407 UP000075840 UP000000437 UP000014760 UP000261540 UP000265040 UP000079169 UP000075901 UP000265000 UP000242638 UP000261360
UP000235965 UP000053825 UP000000311 UP000245037 UP000265140 UP000245119 UP000005203 UP000078540 UP000242457 UP000076502 UP000005205 UP000030742 UP000008672 UP000192224 UP000008820 UP000036403 UP000019118 UP000193380 UP000018467 UP000034805 UP000221080 UP000007755 UP000027135 UP000261560 UP000075809 UP000261440 UP000078492 UP000008237 UP000078200 UP000053105 UP000002320 UP000087266 UP000007266 UP000002358 UP000007110 UP000092460 UP000092443 UP000092444 UP000076420 UP000257200 UP000053454 UP000007303 UP000257160 UP000265080 UP000075883 UP000092445 UP000053097 UP000279307 UP000092461 UP000261400 UP000009046 UP000186698 UP000261580 UP000005207 UP000275408 UP000007062 UP000076407 UP000075840 UP000000437 UP000014760 UP000261540 UP000265040 UP000079169 UP000075901 UP000265000 UP000242638 UP000261360
Pfam
PF03099 BPL_LplA_LipB
Interpro
SUPFAM
SSF53098
SSF53098
CDD
ProteinModelPortal
A0A212FKU4
A0A2A4IZ19
A0A2H1WEA6
A0A194RE29
A0A194PKI5
A0A437BVD1
+ More
A0A0L7LAW1 A0A2J7QYL2 A0A0L7QY58 E2A2V5 A0A2P8XZI4 A0A3P8YKA1 A0A2T7NCM6 A0A088ALW7 A0A151I3F1 A0A2A3E6B0 A0A154NWE4 C1BYA8 A0A158NXY6 U4U015 H3A1P6 A0A1W4YTY4 Q0IEK7 A0A0J7L363 N6T2D6 A0A060WM11 A0A3B1KFN2 A0A0P7UV33 E3TGJ5 F4WMW4 A0A067RKW3 A0A3B3CEV7 A0A3B1JQ22 A0A1S4FN42 A0A151WIG7 A0A3B4DS71 A0A151IVW7 A0A2D0SSE5 A0A0P6FSE3 A0A3N0XI49 E2BQH3 A0A0P5KBG0 A0A1B6LDI8 A0A0P6BGB1 A0A1A9UZU6 A0A0M9A1L5 E9IID1 A0A2R7WBM1 B0WH35 A0A1S3NPT8 D6WCX6 K7IYB8 U5EXR5 W4XML7 A0A1B0AL12 A0A1A9XN22 A0A1B0FR83 A0A2C9JF27 A0A1B6HLI8 A0A3Q1ETW8 A0A437CTR0 A0A0L8FR25 A0A1A8P1U8 Q4SB31 A0A3Q1D0S5 A0A3P8TSF6 A0A182LXS1 A0A1A9ZZQ8 A0A026VU07 A0A1A8J639 A0A1A8C774 A0A1A7ZW42 A0A1B0CL88 A0A131XKJ9 A0A3B5B6N9 E0VS59 A0A1A8H0Q2 A0A1L8HBD3 A0A3Q4N1Z4 I3KKT0 A0A3M6UTA1 A0A0F8BU33 Q7PS31 A0A182XEE9 A0A182IAB4 Q29R99 R7V6C9 A0A1Q3EXD3 A0A1A8L678 A0A2M4CS08 A0A3B3QZL2 A0A3Q1J525 A0A1S3DW12 A0A1L8E0L8 F1QAW5 A0A131YV83 A0A182S5L7 A0A3Q2R0K8 A0A3P9MXY6 A0A3B4ZA36
A0A0L7LAW1 A0A2J7QYL2 A0A0L7QY58 E2A2V5 A0A2P8XZI4 A0A3P8YKA1 A0A2T7NCM6 A0A088ALW7 A0A151I3F1 A0A2A3E6B0 A0A154NWE4 C1BYA8 A0A158NXY6 U4U015 H3A1P6 A0A1W4YTY4 Q0IEK7 A0A0J7L363 N6T2D6 A0A060WM11 A0A3B1KFN2 A0A0P7UV33 E3TGJ5 F4WMW4 A0A067RKW3 A0A3B3CEV7 A0A3B1JQ22 A0A1S4FN42 A0A151WIG7 A0A3B4DS71 A0A151IVW7 A0A2D0SSE5 A0A0P6FSE3 A0A3N0XI49 E2BQH3 A0A0P5KBG0 A0A1B6LDI8 A0A0P6BGB1 A0A1A9UZU6 A0A0M9A1L5 E9IID1 A0A2R7WBM1 B0WH35 A0A1S3NPT8 D6WCX6 K7IYB8 U5EXR5 W4XML7 A0A1B0AL12 A0A1A9XN22 A0A1B0FR83 A0A2C9JF27 A0A1B6HLI8 A0A3Q1ETW8 A0A437CTR0 A0A0L8FR25 A0A1A8P1U8 Q4SB31 A0A3Q1D0S5 A0A3P8TSF6 A0A182LXS1 A0A1A9ZZQ8 A0A026VU07 A0A1A8J639 A0A1A8C774 A0A1A7ZW42 A0A1B0CL88 A0A131XKJ9 A0A3B5B6N9 E0VS59 A0A1A8H0Q2 A0A1L8HBD3 A0A3Q4N1Z4 I3KKT0 A0A3M6UTA1 A0A0F8BU33 Q7PS31 A0A182XEE9 A0A182IAB4 Q29R99 R7V6C9 A0A1Q3EXD3 A0A1A8L678 A0A2M4CS08 A0A3B3QZL2 A0A3Q1J525 A0A1S3DW12 A0A1L8E0L8 F1QAW5 A0A131YV83 A0A182S5L7 A0A3Q2R0K8 A0A3P9MXY6 A0A3B4ZA36
PDB
1W66
E-value=2.53076e-34,
Score=361
Ontologies
PATHWAY
GO
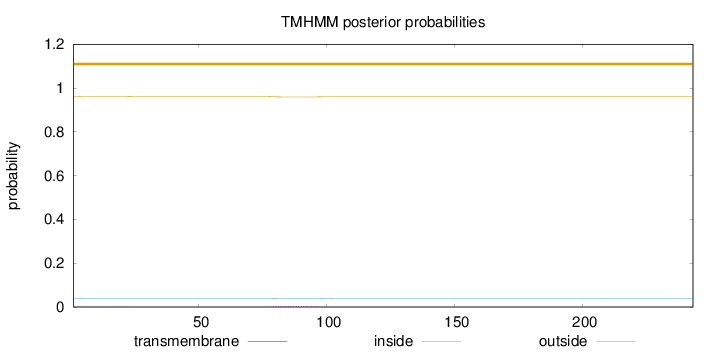
Topology
Subcellular location
Mitochondrion
Length:
243
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.069
Exp number, first 60 AAs:
0.01177
Total prob of N-in:
0.03936
outside
1 - 243
Population Genetic Test Statistics
Pi
11.919336
Theta
18.048248
Tajima's D
-0.86651
CLR
41.541363
CSRT
0.16079196040198
Interpretation
Uncertain
