Gene
KWMTBOMO08879
Annotation
reverse_transcriptase_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.454
Sequence
CDS
ATGGCGCCGCAATTCCAAGGAGTATCCTTGCAAGATTCCGAGAGTATTGGGATACTTGGGGTCGACATTTCGAGCGATGTCCAGTTTCGGAGTCATTTGGAAGGCAAAGCCAAGTTGGCATCCAAAATGCTGGGAGTCCGCAACAGAGCAAAGCGGTACTTCACACCTGGACAAAGGATTTTGCTTTATAAAGCACAAGTCCGGCCTCGCGTGGAGTACTGCTCCCATCTCTGGGCCGGGGCTCCCAAATACCAGCTTATTCCATTTGACTCCATACAGAGGAGGGCCGTTCGGATTGTCGATAATCCCATTCTCACGGATCGTTTGGAACCTCTGGGTCTGCGGAGGGACTTCGGCTCCCTCTGTATTTTGTACCGTATGTTCCATGGGGAGTGCTCTGAGGAATTAATTGTTCGAGATGATACCGACATCTCGTTCTTACCATCGCACCGCCCGTCACCGGAGTAG
Protein
MAPQFQGVSLQDSESIGILGVDISSDVQFRSHLEGKAKLASKMLGVRNRAKRYFTPGQRILLYKAQVRPRVEYCSHLWAGAPKYQLIPFDSIQRRAVRIVDNPILTDRLEPLGLRRDFGSLCILYRMFHGECSEELIVRDDTDISFLPSHRPSPE
Summary
Uniprot
Q5KTM5
Q6UV17
A0A3S2KYN2
A0A437B9X9
A0A3S2LP03
A0A3S2P9Q3
+ More
A0A437ARG3 A0A3S2NTH0 A0A437ARI9 A0A3S2NHP4 A0A3S2LQP1 A0A3S2NMS5 A0A3S2NMX8 A0A437BF96 A0A2H1VFZ3 J9GT67 A0A437BUJ7 A0A2H1WCG9 A0A0P4VXU0 T2M6M7 A0A3D5S1K1 A0A1Y1S3P2 A0A2H1V9A8 A0A437B537 A0A1E1XGQ7 A0A0K8RPQ6 A0A2G8JWN9 A0A147BI04 A0A131XV56 A0A131Y4D1 V5H8T0 W4Y4X9 A0A0P4VPK5 A0A2G8KKJ8 A0A131Y531 A0A0P4VVV8 A0A147BNC9 A0A131Y4Q4 A0A293LEJ4 A0A1E1XM20 A0A1E1XVB3 A0A093HBM3 A0A147BJ69 A0A1B6FN30 A0A1B6GP81 A0A093HQN0 A0A2G8JFI5 A0A093KB34 A0A023FCJ1 A0A147BL51 K1QCH1 A0A093H5S5 A0A093I968 A0A093JFI6 A0A093GU34 A0A093GQT0 A0A147BBR8 A0A093HMV9 V5IE54 A0A093H7K6 A0A093HT80 A0A093HPP7 A0A093HFB7 A0A087QT51 A0A147BRL4
A0A437ARG3 A0A3S2NTH0 A0A437ARI9 A0A3S2NHP4 A0A3S2LQP1 A0A3S2NMS5 A0A3S2NMX8 A0A437BF96 A0A2H1VFZ3 J9GT67 A0A437BUJ7 A0A2H1WCG9 A0A0P4VXU0 T2M6M7 A0A3D5S1K1 A0A1Y1S3P2 A0A2H1V9A8 A0A437B537 A0A1E1XGQ7 A0A0K8RPQ6 A0A2G8JWN9 A0A147BI04 A0A131XV56 A0A131Y4D1 V5H8T0 W4Y4X9 A0A0P4VPK5 A0A2G8KKJ8 A0A131Y531 A0A0P4VVV8 A0A147BNC9 A0A131Y4Q4 A0A293LEJ4 A0A1E1XM20 A0A1E1XVB3 A0A093HBM3 A0A147BJ69 A0A1B6FN30 A0A1B6GP81 A0A093HQN0 A0A2G8JFI5 A0A093KB34 A0A023FCJ1 A0A147BL51 K1QCH1 A0A093H5S5 A0A093I968 A0A093JFI6 A0A093GU34 A0A093GQT0 A0A147BBR8 A0A093HMV9 V5IE54 A0A093H7K6 A0A093HT80 A0A093HPP7 A0A093HFB7 A0A087QT51 A0A147BRL4
Pubmed
EMBL
AB126052
BAD86652.1
AY359886
AAQ57129.1
RSAL01000487
RVE41561.1
+ More
RSAL01000109 RVE47192.1 RSAL01002067 RVE40593.1 RSAL01000164 RVE45343.1 RSAL01002827 RVE40408.1 RSAL01000090 RVE48098.1 RSAL01002868 RVE40398.1 RSAL01003136 RVE40332.1 RSAL01000422 RVE41792.1 RSAL01000002 RVE54942.1 RVE55041.1 RVE55094.1 RSAL01000072 RVE49051.1 ODYU01002162 SOQ39302.1 AMCI01002135 EJX03495.1 RSAL01000007 RVE54134.1 ODYU01007423 SOQ50184.1 GDRN01105059 JAI57791.1 HAAD01001474 CDG67706.1 DPOC01000269 HCX22453.1 LWDP01000391 ORD92879.1 ODYU01001348 SOQ37407.1 RSAL01000157 RVE45592.1 GFAC01000919 JAT98269.1 GADI01001229 JAA72579.1 MRZV01001153 PIK40187.1 GEGO01005479 JAR89925.1 GEFM01006250 JAP69546.1 GEFM01001453 JAP74343.1 GANP01010944 JAB73524.1 AAGJ04110620 GDRN01111278 JAI56803.1 MRZV01000519 PIK48495.1 GEFM01002306 JAP73490.1 GDRN01111277 JAI56804.1 GEGO01003111 JAR92293.1 GEFM01002324 JAP73472.1 GFWV01002422 MAA27152.1 GFAA01003161 JAU00274.1 GFAA01000199 JAU03236.1 KL206225 KFV80018.1 GEGO01004630 JAR90774.1 GECZ01018152 JAS51617.1 GECZ01005534 JAS64235.1 KL206694 KFV84978.1 MRZV01002157 PIK34500.1 KL206955 KFV87539.1 GBBK01004986 JAC19496.1 GEGO01003895 JAR91509.1 JH815916 EKC31648.1 KL206020 KFV77928.1 KL206974 KFV87692.1 KL205964 KFV77614.1 KL205741 KFV73823.1 KL205665 KFV72673.1 GEGO01007187 JAR88217.1 KL206526 KFV83081.1 GANP01010782 JAB73686.1 KL205979 KFV77661.1 KL206805 KFV85878.1 KL206341 KFV81395.1 KL206333 KFV81308.1 KL225891 KFM04405.1 GEGO01002025 JAR93379.1
RSAL01000109 RVE47192.1 RSAL01002067 RVE40593.1 RSAL01000164 RVE45343.1 RSAL01002827 RVE40408.1 RSAL01000090 RVE48098.1 RSAL01002868 RVE40398.1 RSAL01003136 RVE40332.1 RSAL01000422 RVE41792.1 RSAL01000002 RVE54942.1 RVE55041.1 RVE55094.1 RSAL01000072 RVE49051.1 ODYU01002162 SOQ39302.1 AMCI01002135 EJX03495.1 RSAL01000007 RVE54134.1 ODYU01007423 SOQ50184.1 GDRN01105059 JAI57791.1 HAAD01001474 CDG67706.1 DPOC01000269 HCX22453.1 LWDP01000391 ORD92879.1 ODYU01001348 SOQ37407.1 RSAL01000157 RVE45592.1 GFAC01000919 JAT98269.1 GADI01001229 JAA72579.1 MRZV01001153 PIK40187.1 GEGO01005479 JAR89925.1 GEFM01006250 JAP69546.1 GEFM01001453 JAP74343.1 GANP01010944 JAB73524.1 AAGJ04110620 GDRN01111278 JAI56803.1 MRZV01000519 PIK48495.1 GEFM01002306 JAP73490.1 GDRN01111277 JAI56804.1 GEGO01003111 JAR92293.1 GEFM01002324 JAP73472.1 GFWV01002422 MAA27152.1 GFAA01003161 JAU00274.1 GFAA01000199 JAU03236.1 KL206225 KFV80018.1 GEGO01004630 JAR90774.1 GECZ01018152 JAS51617.1 GECZ01005534 JAS64235.1 KL206694 KFV84978.1 MRZV01002157 PIK34500.1 KL206955 KFV87539.1 GBBK01004986 JAC19496.1 GEGO01003895 JAR91509.1 JH815916 EKC31648.1 KL206020 KFV77928.1 KL206974 KFV87692.1 KL205964 KFV77614.1 KL205741 KFV73823.1 KL205665 KFV72673.1 GEGO01007187 JAR88217.1 KL206526 KFV83081.1 GANP01010782 JAB73686.1 KL205979 KFV77661.1 KL206805 KFV85878.1 KL206341 KFV81395.1 KL206333 KFV81308.1 KL225891 KFM04405.1 GEGO01002025 JAR93379.1
Proteomes
Pfam
Interpro
IPR000477
RT_dom
+ More
IPR005135 Endo/exonuclease/phosphatase
IPR036691 Endo/exonu/phosph_ase_sf
IPR016190 Transl_init_fac_IF2/IF5_Zn-bd
IPR002735 Transl_init_fac_IF2/IF5
IPR016189 Transl_init_fac_IF2/IF5_N
IPR007023 Ribosom_reg
IPR028941 WHIM2_dom
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR000504 RRM_dom
IPR034653 SPF45_RRM
IPR003954 RRM_dom_euk
IPR000467 G_patch_dom
IPR040052 RBM17
IPR035979 RBD_domain_sf
IPR036291 NAD(P)-bd_dom_sf
IPR002347 SDR_fam
IPR020904 Sc_DH/Rdtase_CS
IPR036322 WD40_repeat_dom_sf
IPR040382 NOL10/Enp2
IPR012580 NUC153
IPR001680 WD40_repeat
IPR039779 RFX-like
IPR038751 Int8
IPR028082 Peripla_BP_I
IPR005135 Endo/exonuclease/phosphatase
IPR036691 Endo/exonu/phosph_ase_sf
IPR016190 Transl_init_fac_IF2/IF5_Zn-bd
IPR002735 Transl_init_fac_IF2/IF5
IPR016189 Transl_init_fac_IF2/IF5_N
IPR007023 Ribosom_reg
IPR028941 WHIM2_dom
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR000504 RRM_dom
IPR034653 SPF45_RRM
IPR003954 RRM_dom_euk
IPR000467 G_patch_dom
IPR040052 RBM17
IPR035979 RBD_domain_sf
IPR036291 NAD(P)-bd_dom_sf
IPR002347 SDR_fam
IPR020904 Sc_DH/Rdtase_CS
IPR036322 WD40_repeat_dom_sf
IPR040382 NOL10/Enp2
IPR012580 NUC153
IPR001680 WD40_repeat
IPR039779 RFX-like
IPR038751 Int8
IPR028082 Peripla_BP_I
SUPFAM
Gene 3D
ProteinModelPortal
Q5KTM5
Q6UV17
A0A3S2KYN2
A0A437B9X9
A0A3S2LP03
A0A3S2P9Q3
+ More
A0A437ARG3 A0A3S2NTH0 A0A437ARI9 A0A3S2NHP4 A0A3S2LQP1 A0A3S2NMS5 A0A3S2NMX8 A0A437BF96 A0A2H1VFZ3 J9GT67 A0A437BUJ7 A0A2H1WCG9 A0A0P4VXU0 T2M6M7 A0A3D5S1K1 A0A1Y1S3P2 A0A2H1V9A8 A0A437B537 A0A1E1XGQ7 A0A0K8RPQ6 A0A2G8JWN9 A0A147BI04 A0A131XV56 A0A131Y4D1 V5H8T0 W4Y4X9 A0A0P4VPK5 A0A2G8KKJ8 A0A131Y531 A0A0P4VVV8 A0A147BNC9 A0A131Y4Q4 A0A293LEJ4 A0A1E1XM20 A0A1E1XVB3 A0A093HBM3 A0A147BJ69 A0A1B6FN30 A0A1B6GP81 A0A093HQN0 A0A2G8JFI5 A0A093KB34 A0A023FCJ1 A0A147BL51 K1QCH1 A0A093H5S5 A0A093I968 A0A093JFI6 A0A093GU34 A0A093GQT0 A0A147BBR8 A0A093HMV9 V5IE54 A0A093H7K6 A0A093HT80 A0A093HPP7 A0A093HFB7 A0A087QT51 A0A147BRL4
A0A437ARG3 A0A3S2NTH0 A0A437ARI9 A0A3S2NHP4 A0A3S2LQP1 A0A3S2NMS5 A0A3S2NMX8 A0A437BF96 A0A2H1VFZ3 J9GT67 A0A437BUJ7 A0A2H1WCG9 A0A0P4VXU0 T2M6M7 A0A3D5S1K1 A0A1Y1S3P2 A0A2H1V9A8 A0A437B537 A0A1E1XGQ7 A0A0K8RPQ6 A0A2G8JWN9 A0A147BI04 A0A131XV56 A0A131Y4D1 V5H8T0 W4Y4X9 A0A0P4VPK5 A0A2G8KKJ8 A0A131Y531 A0A0P4VVV8 A0A147BNC9 A0A131Y4Q4 A0A293LEJ4 A0A1E1XM20 A0A1E1XVB3 A0A093HBM3 A0A147BJ69 A0A1B6FN30 A0A1B6GP81 A0A093HQN0 A0A2G8JFI5 A0A093KB34 A0A023FCJ1 A0A147BL51 K1QCH1 A0A093H5S5 A0A093I968 A0A093JFI6 A0A093GU34 A0A093GQT0 A0A147BBR8 A0A093HMV9 V5IE54 A0A093H7K6 A0A093HT80 A0A093HPP7 A0A093HFB7 A0A087QT51 A0A147BRL4
Ontologies
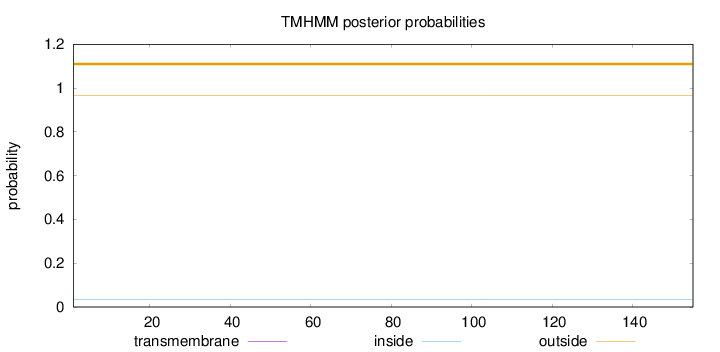
Topology
Length:
155
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00019
Exp number, first 60 AAs:
0.00019
Total prob of N-in:
0.03551
outside
1 - 155
Population Genetic Test Statistics
Pi
115.973449
Theta
106.069009
Tajima's D
-0.353239
CLR
0
CSRT
0.281685915704215
Interpretation
Uncertain
