Gene
KWMTBOMO08877
Annotation
reverse_transcriptase_[Bombyx_mori]
Full name
Probable RNA-directed DNA polymerase from transposon BS
Alternative Name
Reverse transcriptase
Location in the cell
Nuclear Reliability : 2.736
Sequence
CDS
ATGCGTGCCCTACAACGCGACGTAAAGTCTCGCATCGCCGAAGTCCGAGATGCCAGATGGTCTGATTTCTTAGAAGGACTCGCGCCCTCCCAAAGGTCTTACTACCGCTTAGCTCGTACTCTCAAATCGGATACGGTAGTAACTATGCCCCCCCTCGTAGGCCCCTCAGGCCGACTCGCGGCGTTCGATGATGACGAAAAAGCAGAGCTGCTGGCCGATACATTGCAAACCCAGTGCACGCCCAGCACTCAATCCGTGGACCCTGTTCATGTAGAATTAGTAGACAGTGAGGTAGAACGCAGAGCCTCCTTGCCACCATCGGATGCGTTACCACCCGTCACCCCGATAGAAGTTAAAGACTTGATCAAAGACCTACGTCCTCGCAAGGCTCCCGGTTCCGACTGTATATCTAACCGCGTTATTAAACTTCTACCCGTCCAACTCATCGTGATGTTGGCATCTATTTTCAATGCCGCTATGGCGAACTGTATCTTTCCCGCGGTGTGGAAAGAAGCGGACGTTATCGGCATACATAAACCCGGTAAACCAAAAAATCATCCGACGAGCTACCGCCCGATTAGCCTCCTCATGTCTCTAGGCAAACTGTATGAGCGTCTGCTCTACAAACGCCTCAGAGACTTCGTCTCATCCAAGGGCATTCTTATCGATGAACAATTCGGATTCCGTACAAATCACTCATGCGTTCAACAGGTGCACCGCCTCTATAAAAACACAACTATATTCTGTGTACAGCAGTCTGGCTTAACACGTGTCCCTAGTGAGCTAGTTTTGTTACATCTGAAAAGTTGTTTAGGGCGACTTAGAGAGGTTCAGTTAGGCGAGTATGTAATCACGAGTTTAACGGTGTTAGAAATTCTCAGGTTCATTAAATTATAA
Protein
MRALQRDVKSRIAEVRDARWSDFLEGLAPSQRSYYRLARTLKSDTVVTMPPLVGPSGRLAAFDDDEKAELLADTLQTQCTPSTQSVDPVHVELVDSEVERRASLPPSDALPPVTPIEVKDLIKDLRPRKAPGSDCISNRVIKLLPVQLIVMLASIFNAAMANCIFPAVWKEADVIGIHKPGKPKNHPTSYRPISLLMSLGKLYERLLYKRLRDFVSSKGILIDEQFGFRTNHSCVQQVHRLYKNTTIFCVQQSGLTRVPSELVLLHLKSCLGRLREVQLGEYVITSLTVLEILRFIKL
Summary
Catalytic Activity
a 2'-deoxyribonucleoside 5'-triphosphate + DNA(n) = diphosphate + DNA(n+1)
Cofactor
Mg(2+)
Mn(2+)
Mn(2+)
Keywords
Nucleotidyltransferase
RNA-directed DNA polymerase
Transferase
Transposable element
Feature
chain Probable RNA-directed DNA polymerase from transposon BS
Uniprot
Q93137
Q9XXW0
Q9BPQ0
A0A0N1PFV1
O96546
D7EM17
+ More
A0A139W8G9 A0A1Y1NA23 A0A139WA11 A0A2S2Q352 A0A1B6J5Z9 A0A2S2NPT6 U4U0U7 V5GNJ2 J9KLB1 A0A2H8U0H5 A0A023F6S4 A0A2S2PEZ9 A0A2S2QEE0 A0A0C9RJA4 V5GNF3 X1WVL6 A0A224X6S1 A0A087UDC5 K7IN67 X1WKS2 A0A2S2PA10 A0A2S2N6C8 A0A087UCW3 A0A2S2N629 A0A3N5Z7I3 A0A2J7R7D0 A0A2S2NCK2 A0A1Y1K0C8 A0A1Y1LKN6 A0A224X6U5 A0A224X7Z3 Q95SX7 A0A1Y1KFT3 A0A2J7Q0S7 A0A2S2N814 A0A224XI99 A0A1Y1KFR3 A0A2S2PWA8 A0A2S2Q5V3 J9K7I4 A0A1B6C2V8 A0A023F700 A0A2S2PPK8 A0A2S2RB48 A0A2H8TKI9 A0A2S2NH02 A0A023F754 A0A2S2QLY2 A0A2S2PBB4 Q07995 A0A2S2PR14 A0A069DZC5 A0A069DW01 J9KKL3 A0A1Y1MHM1 A0A224X6T4 A0A087UB64 A0A069DVA6 A0A023F069 A0A2S2R9D4 A0A2S2P3X8 A0A224X6W2 A0A023EYS8 A0A224X724
A0A139W8G9 A0A1Y1NA23 A0A139WA11 A0A2S2Q352 A0A1B6J5Z9 A0A2S2NPT6 U4U0U7 V5GNJ2 J9KLB1 A0A2H8U0H5 A0A023F6S4 A0A2S2PEZ9 A0A2S2QEE0 A0A0C9RJA4 V5GNF3 X1WVL6 A0A224X6S1 A0A087UDC5 K7IN67 X1WKS2 A0A2S2PA10 A0A2S2N6C8 A0A087UCW3 A0A2S2N629 A0A3N5Z7I3 A0A2J7R7D0 A0A2S2NCK2 A0A1Y1K0C8 A0A1Y1LKN6 A0A224X6U5 A0A224X7Z3 Q95SX7 A0A1Y1KFT3 A0A2J7Q0S7 A0A2S2N814 A0A224XI99 A0A1Y1KFR3 A0A2S2PWA8 A0A2S2Q5V3 J9K7I4 A0A1B6C2V8 A0A023F700 A0A2S2PPK8 A0A2S2RB48 A0A2H8TKI9 A0A2S2NH02 A0A023F754 A0A2S2QLY2 A0A2S2PBB4 Q07995 A0A2S2PR14 A0A069DZC5 A0A069DW01 J9KKL3 A0A1Y1MHM1 A0A224X6T4 A0A087UB64 A0A069DVA6 A0A023F069 A0A2S2R9D4 A0A2S2P3X8 A0A224X6W2 A0A023EYS8 A0A224X724
EC Number
2.7.7.49
Pubmed
EMBL
U07847
AAA17752.1
AB018558
BAA76304.1
AB055391
BAB21761.1
+ More
KQ459265 KPJ02064.1 AF081103 AAC72921.1 KQ973412 EFA12510.2 KQ973342 KXZ75569.1 GEZM01008457 JAV94772.1 KQ971729 KYB24751.1 GGMS01002955 MBY72158.1 GECU01013097 JAS94609.1 GGMR01006581 MBY19200.1 KB631961 ERL87484.1 GALX01005299 JAB63167.1 ABLF02040174 ABLF02040183 ABLF02055266 GFXV01007093 MBW18898.1 GBBI01001646 JAC17066.1 GGMR01015412 MBY28031.1 GGMS01006359 MBY75562.1 GBYB01008290 JAG78057.1 GALX01005359 JAB63107.1 ABLF02042385 GFTR01008271 JAW08155.1 KK119316 KFM75364.1 ABLF02005245 ABLF02013477 ABLF02047385 ABLF02066435 GGMR01013409 MBY26028.1 GGMR01000086 MBY12705.1 KK119261 KFM75202.1 GGMR01000005 MBY12624.1 RPOV01000134 RPJ78669.1 NEVH01006736 PNF36716.1 GGMR01002322 MBY14941.1 GEZM01099921 GEZM01099920 GEZM01099918 GEZM01099906 JAV53125.1 GEZM01053022 JAV74219.1 GFTR01008251 JAW08175.1 GFTR01007929 JAW08497.1 AY060438 AAL25477.1 GEZM01084970 JAV60359.1 NEVH01019963 PNF22185.1 GGMR01000676 MBY13295.1 GFTR01008256 JAW08170.1 GEZM01084969 JAV60363.1 GGMS01000625 MBY69828.1 GGMS01003914 MBY73117.1 ABLF02017543 ABLF02017549 ABLF02017551 ABLF02021996 GEDC01029594 JAS07704.1 GBBI01001699 JAC17013.1 GGMR01018716 MBY31335.1 GGMS01018022 MBY87225.1 GFXV01002840 MBW14645.1 GGMR01003864 MBY16483.1 GBBI01001642 JAC17070.1 GGMS01009480 MBY78683.1 GGMR01014053 MBY26672.1 S59870 AAB26437.2 GGMR01019198 MBY31817.1 GBGD01000880 JAC88009.1 GBGD01000799 JAC88090.1 ABLF02029306 ABLF02029314 ABLF02060138 GEZM01032972 JAV84478.1 GFTR01008261 JAW08165.1 KK119069 KFM74603.1 GBGD01000881 JAC88008.1 GBBI01003810 JAC14902.1 GGMS01017365 MBY86568.1 GGMR01011511 MBY24130.1 GFTR01008260 JAW08166.1 GBBI01004395 JAC14317.1 GFTR01008169 JAW08257.1
KQ459265 KPJ02064.1 AF081103 AAC72921.1 KQ973412 EFA12510.2 KQ973342 KXZ75569.1 GEZM01008457 JAV94772.1 KQ971729 KYB24751.1 GGMS01002955 MBY72158.1 GECU01013097 JAS94609.1 GGMR01006581 MBY19200.1 KB631961 ERL87484.1 GALX01005299 JAB63167.1 ABLF02040174 ABLF02040183 ABLF02055266 GFXV01007093 MBW18898.1 GBBI01001646 JAC17066.1 GGMR01015412 MBY28031.1 GGMS01006359 MBY75562.1 GBYB01008290 JAG78057.1 GALX01005359 JAB63107.1 ABLF02042385 GFTR01008271 JAW08155.1 KK119316 KFM75364.1 ABLF02005245 ABLF02013477 ABLF02047385 ABLF02066435 GGMR01013409 MBY26028.1 GGMR01000086 MBY12705.1 KK119261 KFM75202.1 GGMR01000005 MBY12624.1 RPOV01000134 RPJ78669.1 NEVH01006736 PNF36716.1 GGMR01002322 MBY14941.1 GEZM01099921 GEZM01099920 GEZM01099918 GEZM01099906 JAV53125.1 GEZM01053022 JAV74219.1 GFTR01008251 JAW08175.1 GFTR01007929 JAW08497.1 AY060438 AAL25477.1 GEZM01084970 JAV60359.1 NEVH01019963 PNF22185.1 GGMR01000676 MBY13295.1 GFTR01008256 JAW08170.1 GEZM01084969 JAV60363.1 GGMS01000625 MBY69828.1 GGMS01003914 MBY73117.1 ABLF02017543 ABLF02017549 ABLF02017551 ABLF02021996 GEDC01029594 JAS07704.1 GBBI01001699 JAC17013.1 GGMR01018716 MBY31335.1 GGMS01018022 MBY87225.1 GFXV01002840 MBW14645.1 GGMR01003864 MBY16483.1 GBBI01001642 JAC17070.1 GGMS01009480 MBY78683.1 GGMR01014053 MBY26672.1 S59870 AAB26437.2 GGMR01019198 MBY31817.1 GBGD01000880 JAC88009.1 GBGD01000799 JAC88090.1 ABLF02029306 ABLF02029314 ABLF02060138 GEZM01032972 JAV84478.1 GFTR01008261 JAW08165.1 KK119069 KFM74603.1 GBGD01000881 JAC88008.1 GBBI01003810 JAC14902.1 GGMS01017365 MBY86568.1 GGMR01011511 MBY24130.1 GFTR01008260 JAW08166.1 GBBI01004395 JAC14317.1 GFTR01008169 JAW08257.1
Proteomes
Pfam
Interpro
IPR036691
Endo/exonu/phosph_ase_sf
+ More
IPR005135 Endo/exonuclease/phosphatase
IPR000477 RT_dom
IPR006579 Pre_C2HC_dom
IPR013103 RVT_2
IPR041588 Integrase_H2C2
IPR036397 RNaseH_sf
IPR005312 DUF1759
IPR040676 DUF5641
IPR008737 Peptidase_asp_put
IPR008042 Retrotrans_Pao
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
IPR005135 Endo/exonuclease/phosphatase
IPR000477 RT_dom
IPR006579 Pre_C2HC_dom
IPR013103 RVT_2
IPR041588 Integrase_H2C2
IPR036397 RNaseH_sf
IPR005312 DUF1759
IPR040676 DUF5641
IPR008737 Peptidase_asp_put
IPR008042 Retrotrans_Pao
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
Gene 3D
ProteinModelPortal
Q93137
Q9XXW0
Q9BPQ0
A0A0N1PFV1
O96546
D7EM17
+ More
A0A139W8G9 A0A1Y1NA23 A0A139WA11 A0A2S2Q352 A0A1B6J5Z9 A0A2S2NPT6 U4U0U7 V5GNJ2 J9KLB1 A0A2H8U0H5 A0A023F6S4 A0A2S2PEZ9 A0A2S2QEE0 A0A0C9RJA4 V5GNF3 X1WVL6 A0A224X6S1 A0A087UDC5 K7IN67 X1WKS2 A0A2S2PA10 A0A2S2N6C8 A0A087UCW3 A0A2S2N629 A0A3N5Z7I3 A0A2J7R7D0 A0A2S2NCK2 A0A1Y1K0C8 A0A1Y1LKN6 A0A224X6U5 A0A224X7Z3 Q95SX7 A0A1Y1KFT3 A0A2J7Q0S7 A0A2S2N814 A0A224XI99 A0A1Y1KFR3 A0A2S2PWA8 A0A2S2Q5V3 J9K7I4 A0A1B6C2V8 A0A023F700 A0A2S2PPK8 A0A2S2RB48 A0A2H8TKI9 A0A2S2NH02 A0A023F754 A0A2S2QLY2 A0A2S2PBB4 Q07995 A0A2S2PR14 A0A069DZC5 A0A069DW01 J9KKL3 A0A1Y1MHM1 A0A224X6T4 A0A087UB64 A0A069DVA6 A0A023F069 A0A2S2R9D4 A0A2S2P3X8 A0A224X6W2 A0A023EYS8 A0A224X724
A0A139W8G9 A0A1Y1NA23 A0A139WA11 A0A2S2Q352 A0A1B6J5Z9 A0A2S2NPT6 U4U0U7 V5GNJ2 J9KLB1 A0A2H8U0H5 A0A023F6S4 A0A2S2PEZ9 A0A2S2QEE0 A0A0C9RJA4 V5GNF3 X1WVL6 A0A224X6S1 A0A087UDC5 K7IN67 X1WKS2 A0A2S2PA10 A0A2S2N6C8 A0A087UCW3 A0A2S2N629 A0A3N5Z7I3 A0A2J7R7D0 A0A2S2NCK2 A0A1Y1K0C8 A0A1Y1LKN6 A0A224X6U5 A0A224X7Z3 Q95SX7 A0A1Y1KFT3 A0A2J7Q0S7 A0A2S2N814 A0A224XI99 A0A1Y1KFR3 A0A2S2PWA8 A0A2S2Q5V3 J9K7I4 A0A1B6C2V8 A0A023F700 A0A2S2PPK8 A0A2S2RB48 A0A2H8TKI9 A0A2S2NH02 A0A023F754 A0A2S2QLY2 A0A2S2PBB4 Q07995 A0A2S2PR14 A0A069DZC5 A0A069DW01 J9KKL3 A0A1Y1MHM1 A0A224X6T4 A0A087UB64 A0A069DVA6 A0A023F069 A0A2S2R9D4 A0A2S2P3X8 A0A224X6W2 A0A023EYS8 A0A224X724
Ontologies
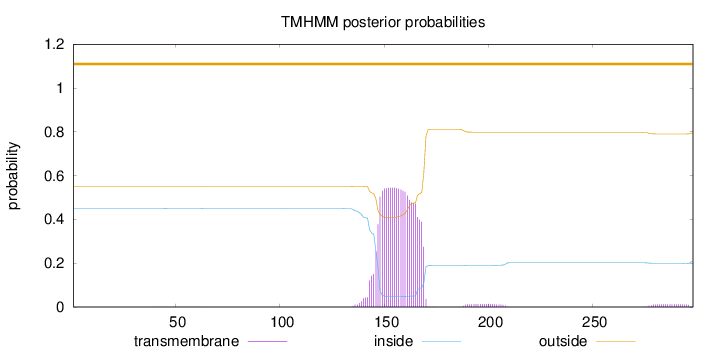
Topology
Length:
298
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
12.62355
Exp number, first 60 AAs:
0.00861
Total prob of N-in:
0.44974
outside
1 - 298
Population Genetic Test Statistics
Pi
6.859009
Theta
9.017322
Tajima's D
-0.697483
CLR
0.042144
CSRT
0.19974001299935
Interpretation
Uncertain
