Gene
KWMTBOMO08875
Pre Gene Modal
BGIBMGA007837
Annotation
PREDICTED:_gamma-aminobutyric_acid_type_B_receptor_subunit_1_isoform_X2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 3.896
Sequence
CDS
ATGTTTCTGCTATGGTTGAGCGCTTGTGGGGGCTATAGTGAAGGAAGACGCGACAATGACCTGCATATCGGCGGCATTTTCCCTATGGAGGGTGAAGGTGGCTGGCAGGGAGGACAAGCGTGTATGCCAGCCGCAGAACTCGCGCTTGCCGACGTGAATGCCAGATCGGACCTATTATCCGGATTTAAGCTACGTCTGCACAGCAACGACAGCAAGTGCGAACCAGGGTTGGGAGCCAGCGTCATGTACAATTTACTGTATAACCCACCGCAAAAATTGTTGCTGCTAGCAGGATGTTCGACAGTTTGTACTACAGTCGCGGAAGCTGCTAAGATGTGGAACCTAATGGTCTTGTGCTATGGAGCTTCGTCGCCAGCGCTGTCGGATCGGTCTCGTTTCCCGACGTTATTCCGAACGCACCCGTCAGCAACAGTCCACAACCCGACGAGAATCAAGCTGATGCAGAAGTTCGCCTGGTCACGAATCGCTATACTGCAACAGGCCGAGGAAGTTTTTATTTCGACTGTCGAAGATTTGGAGGCTCACTGCAAGAAGGCCGGGATCGAAATCGTAACCCGCCAATCGTTTCTATCAGACCCAGCAGACGCCGTGAGGAATCTTCGTCGTCAAGATGCGAGAGTGATCGTCGGCCTGTTCTACGTAGTAGCAGCCAGGAGAGTACTCTGTGAAGTATACAAGCATAGACTATACGGAAAATCCTACGTGTGGTTCTTTATAGGTTGGTATGAGGATAATTGGTTCGAGACGAATCTAGAACGGGAAGGCATCGAATGCACGGTGGAACAGATGAGAGAAGCGGCCGAAGGTCACCTCACAACCGAAGCTCTAATGTGGAATCAAAATGCCAACCAAACTACGATATCCGGAATGACATCTGAAGATTTTAGGTCAAGATTAAATGAGGCACTCCGCGAAGCTGGCTACGACATCGATGGAGAACGGTATCCGGAAGGCTACCAAGAGGCACCCTTGGCTTATGATGCAGTCTGGTCTGTTGCTCTGGCTTTCAATAAAACAATGGAGAAACTGCAGAAGAGCGGTTTGAGTTTGAAGAATTTCACGTACACAAACAAGAAAATAGCTGATGACATTTACGAAGCCATAAATTCGACGTCGTTCCTTGGTGTTTCGGGATTGGTAGCATTTTCGTCTCAAGGTGATCGCATAGCTCTGACACAAATCGAACAATTAACAGACAATCATTACGTGAAACTTGGATATTATGATACACAAGCAGACAACCTTACTTGGCTAGATCGGGAACGGTGGGTCGGTGGAAAAGTCCCTCCCGATCGAACTGTGGTCGTCAATGAGCTGCGAACGGTTTCTCTGCCGCTTTTCGCGTGTATGACGGCCCTCTGCATCGCTGGAATATTGGCAGCCATAGCTCTTATCGTATTTAATATAATGCATAGACACAGAAGAGTAATACAATGGTCCCACCCGGCTTGCAACACCGTAATGCTTTGTGGATGCTGCGTGTGTCTCTGTGCGGCAGTAGCTTTGGGCGTCGATGGGCGATGGGTACTACCTCAACACTTCAATGGATTGTGCGCTGCCCGAGCTTGGCTCTTGGCTACAGGATTCTCGATGGCATATGGAGCAATGTTTACGAAAGTCTGGAGGGTACATCGACATACGACTAAACCAAAAATTGATAATAAGAAACGCATTCATGGTTGGAAACTGTATACAATGGTCGGGGGACTGCTCGTTGTAGACGTTGCACTCCTGACAGCTTGGCAGCTCCGGGACCCGCTGCAGCGACGCGTCGAGACATTCCCCCTGGAAGCCCCGCGACATCACGACGACGACGTGCACATACGACCCGAATTGGAACACTGTGAGAGCAAACATAACACCGTGTGGCTTGGCGTAATGTACGGGTACAAAGGCCTGGTGCTAGTTTTCGGACTGTTCCTGGCCTACGAGACACGCTCGGTGAAGGTTCGACAGATTAATGACTCTCGCTACGTCGGCATGAGCATATACAACGTGGTCGTGTTGTGCCTTATCACTGCGCCGGTTACGTTGGTCATTGCGAGTCAACAAGACGCAGCGTTCGCCTTCGTTTCCCTAGCGATTGTCTTCTGTTGCTTCCTGAGTATGGCCCTAATATTCGTACCTAAGGTAATAGAAGTGATTCGTCATCCAACCGAGCGCGTGGAGAGCGGTCGAGGGTCTGGTTCCGGCTCCGCGCCGGCCGACGAGGAACGCTACCGGGAACTAGTCAAAGAAAATGAAGAATTACAAAAATTAATTACACAAGTACGTACGCTTATGAACGAACTAATAACACGGGCGTTACCGGTCTTCTATATCTTTGTACTCTAA
Protein
MFLLWLSACGGYSEGRRDNDLHIGGIFPMEGEGGWQGGQACMPAAELALADVNARSDLLSGFKLRLHSNDSKCEPGLGASVMYNLLYNPPQKLLLLAGCSTVCTTVAEAAKMWNLMVLCYGASSPALSDRSRFPTLFRTHPSATVHNPTRIKLMQKFAWSRIAILQQAEEVFISTVEDLEAHCKKAGIEIVTRQSFLSDPADAVRNLRRQDARVIVGLFYVVAARRVLCEVYKHRLYGKSYVWFFIGWYEDNWFETNLEREGIECTVEQMREAAEGHLTTEALMWNQNANQTTISGMTSEDFRSRLNEALREAGYDIDGERYPEGYQEAPLAYDAVWSVALAFNKTMEKLQKSGLSLKNFTYTNKKIADDIYEAINSTSFLGVSGLVAFSSQGDRIALTQIEQLTDNHYVKLGYYDTQADNLTWLDRERWVGGKVPPDRTVVVNELRTVSLPLFACMTALCIAGILAAIALIVFNIMHRHRRVIQWSHPACNTVMLCGCCVCLCAAVALGVDGRWVLPQHFNGLCAARAWLLATGFSMAYGAMFTKVWRVHRHTTKPKIDNKKRIHGWKLYTMVGGLLVVDVALLTAWQLRDPLQRRVETFPLEAPRHHDDDVHIRPELEHCESKHNTVWLGVMYGYKGLVLVFGLFLAYETRSVKVRQINDSRYVGMSIYNVVVLCLITAPVTLVIASQQDAAFAFVSLAIVFCCFLSMALIFVPKVIEVIRHPTERVESGRGSGSGSAPADEERYRELVKENEELQKLITQVRTLMNELITRALPVFYIFVL
Summary
Similarity
Belongs to the G-protein coupled receptor 3 family.
Uniprot
H9JE90
A0A3S2M9U9
S6DR08
A0A2H1WAC6
A0A194RD58
A0A2A4JSW0
+ More
A0A194PQ25 A0A310SM81 B3MLC7 A0A0L0BVC8 B4JAW8 A0A0L7R583 A0A0R1DST3 A0A0R1DQW3 A0A0R1DSQ2 Q9V3Q9 B4P0X4 B4MZ77 A0A0R1DLG3 B3N5N0 B4LRY4 B4KLC2 A0A0Q5WMT4 W8BH72 A0A3B0KBC3 A0A0R3NQZ1 A0A0Q5WIS6 A0A1W4W038 Q29P68 Q9BML7 A0A0Q5W991 B4HXL4 A0A1S4H3I9 A0A182I0W8 T1I7D6 A0A182KJX6 A0A0J9R2J4 A0A182YN14 A0A182PPU0 A0A182JP69 A0A182Q9X6 A0A182N8X1 V6DNG0 Q7PME5 A0A182RAF4 A0A1B0CTX3 K7IRD3 A0A2J7Q0I1 W5JI42 A0A182F8W2 A0A088ACG8 A0A084W5M4 A0A2A3EH29 A0A026WP30 A0A232F0L8 A0A3L8E2F5 E1ZXC0 A0A195BEF8 A0A158NQP1 A0A151XB41 F4WBW8 A0A182VSS1 A0A182MVL9 A0A0K8TG24 E0VWF6 A0A195FSC0 A0A0M9AA59 A0A1B6IMJ1 A0A067RJX4 A0A1I8MUN8 E2BM97 B4GK77 B4Q5Y9 A0A1J1IUN2 A0A1S4FG88 A0A195D3M2 A0A2H8TER2 A0A1I8PIE9 E9J436 A0A1Y1LWI4 A0A1J0FAU9 A0A182V1I8 A0A182U5X5 A0A182XCP2 A0A1J0FAT2 A0A1D2NC33 A0A226E705 A0A0P5LWC8 A0A0P6GYC0 A0A182J9B7 A0A0P5TKA2 A0A0P5BNQ1 A0A0P6C3F4 E9G2I1 N6UCG3 A0A061QLL2 A0A1E1X357 H9B3X8 A0A2L2Y1Y5 A0A147BN21
A0A194PQ25 A0A310SM81 B3MLC7 A0A0L0BVC8 B4JAW8 A0A0L7R583 A0A0R1DST3 A0A0R1DQW3 A0A0R1DSQ2 Q9V3Q9 B4P0X4 B4MZ77 A0A0R1DLG3 B3N5N0 B4LRY4 B4KLC2 A0A0Q5WMT4 W8BH72 A0A3B0KBC3 A0A0R3NQZ1 A0A0Q5WIS6 A0A1W4W038 Q29P68 Q9BML7 A0A0Q5W991 B4HXL4 A0A1S4H3I9 A0A182I0W8 T1I7D6 A0A182KJX6 A0A0J9R2J4 A0A182YN14 A0A182PPU0 A0A182JP69 A0A182Q9X6 A0A182N8X1 V6DNG0 Q7PME5 A0A182RAF4 A0A1B0CTX3 K7IRD3 A0A2J7Q0I1 W5JI42 A0A182F8W2 A0A088ACG8 A0A084W5M4 A0A2A3EH29 A0A026WP30 A0A232F0L8 A0A3L8E2F5 E1ZXC0 A0A195BEF8 A0A158NQP1 A0A151XB41 F4WBW8 A0A182VSS1 A0A182MVL9 A0A0K8TG24 E0VWF6 A0A195FSC0 A0A0M9AA59 A0A1B6IMJ1 A0A067RJX4 A0A1I8MUN8 E2BM97 B4GK77 B4Q5Y9 A0A1J1IUN2 A0A1S4FG88 A0A195D3M2 A0A2H8TER2 A0A1I8PIE9 E9J436 A0A1Y1LWI4 A0A1J0FAU9 A0A182V1I8 A0A182U5X5 A0A182XCP2 A0A1J0FAT2 A0A1D2NC33 A0A226E705 A0A0P5LWC8 A0A0P6GYC0 A0A182J9B7 A0A0P5TKA2 A0A0P5BNQ1 A0A0P6C3F4 E9G2I1 N6UCG3 A0A061QLL2 A0A1E1X357 H9B3X8 A0A2L2Y1Y5 A0A147BN21
Pubmed
19121390
26354079
17994087
26108605
17550304
10731132
+ More
12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 15632085 11168554 12364791 20966253 22936249 25244985 20075255 20920257 23761445 24438588 24508170 28648823 30249741 20798317 21347285 21719571 20566863 24845553 25315136 17510324 21282665 28004739 27289101 21292972 23537049 28503490 22403662 26561354 29652888
12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 15632085 11168554 12364791 20966253 22936249 25244985 20075255 20920257 23761445 24438588 24508170 28648823 30249741 20798317 21347285 21719571 20566863 24845553 25315136 17510324 21282665 28004739 27289101 21292972 23537049 28503490 22403662 26561354 29652888
EMBL
BABH01002757
BABH01002758
BABH01002759
BABH01002760
RSAL01000005
RVE54351.1
+ More
HG004164 CDF56917.1 ODYU01007216 SOQ49802.1 KQ460367 KPJ15547.1 NWSH01000638 PCG75101.1 KQ459602 KPI93235.1 KQ759886 OAD62141.1 CH902620 EDV30716.1 JRES01001277 KNC24020.1 CH916368 EDW02838.1 KQ414654 KOC66008.1 CM000157 KRJ98145.1 KRJ98146.1 KRJ98144.1 AE014134 BT031099 AAF53431.3 ABX00721.1 AFH03707.1 EDW89048.2 CH963913 EDW77473.2 KRJ98143.1 KRJ98147.1 CH954177 EDV57989.2 CH940649 EDW63660.2 CH933807 EDW11783.2 KQS70216.1 GAMC01010222 GAMC01010221 JAB96333.1 OUUW01000046 SPP90012.1 CH379058 KRT03495.1 KQS70217.1 EAL34425.2 AF318272 AAK13420.1 KQS70215.1 CH480818 EDW51794.1 AAAB01008980 APCN01005282 ACPB03006256 CM002910 KMY90316.1 AXCN02001871 HG794357 CDK37791.1 EAA13942.4 AJWK01028079 AJWK01028080 NEVH01019967 PNF22098.1 ADMH02001341 ETN62953.1 ATLV01020674 KE525304 KFB45518.1 KZ288256 PBC30592.1 KK107154 EZA56859.1 NNAY01001402 OXU24099.1 QOIP01000001 RLU26555.1 GL435030 EFN74210.1 KQ976511 KYM82550.1 ADTU01023455 KQ982335 KYQ57564.1 GL888066 EGI68396.1 AXCM01000163 GBRD01001460 GBRD01001459 JAG64361.1 DS235819 EEB17707.1 KQ981280 KYN43485.1 KQ435702 KOX80340.1 GECU01019577 JAS88129.1 KK852463 KDR23323.1 GL449158 EFN83223.1 CH479184 EDW37043.1 CM000361 EDX05092.1 CVRI01000058 CRL02846.1 KQ976885 KYN07498.1 GFXV01000766 MBW12571.1 GL768069 EFZ12431.1 GEZM01047923 JAV76690.1 KU986874 APC26103.1 KU986868 APC26097.1 LJIJ01000095 ODN02817.1 LNIX01000007 OXA52376.1 GDIQ01177444 GDIQ01173250 GDIQ01166409 GDIQ01162156 GDIP01044550 LRGB01000996 JAK78475.1 JAM59165.1 KZS14238.1 GDIQ01027552 JAN67185.1 GDIP01125131 JAL78583.1 GDIP01182023 JAJ41379.1 GDIP01009869 JAM93846.1 GL732530 EFX86255.1 APGK01040537 APGK01040538 APGK01040539 APGK01040540 APGK01040541 APGK01040542 KB740983 ENN76342.1 GBFC01000039 JAC59299.1 GFAC01005516 JAT93672.1 JN974907 AFC88976.1 IAAA01002824 LAA02196.1 GEGO01003217 JAR92187.1
HG004164 CDF56917.1 ODYU01007216 SOQ49802.1 KQ460367 KPJ15547.1 NWSH01000638 PCG75101.1 KQ459602 KPI93235.1 KQ759886 OAD62141.1 CH902620 EDV30716.1 JRES01001277 KNC24020.1 CH916368 EDW02838.1 KQ414654 KOC66008.1 CM000157 KRJ98145.1 KRJ98146.1 KRJ98144.1 AE014134 BT031099 AAF53431.3 ABX00721.1 AFH03707.1 EDW89048.2 CH963913 EDW77473.2 KRJ98143.1 KRJ98147.1 CH954177 EDV57989.2 CH940649 EDW63660.2 CH933807 EDW11783.2 KQS70216.1 GAMC01010222 GAMC01010221 JAB96333.1 OUUW01000046 SPP90012.1 CH379058 KRT03495.1 KQS70217.1 EAL34425.2 AF318272 AAK13420.1 KQS70215.1 CH480818 EDW51794.1 AAAB01008980 APCN01005282 ACPB03006256 CM002910 KMY90316.1 AXCN02001871 HG794357 CDK37791.1 EAA13942.4 AJWK01028079 AJWK01028080 NEVH01019967 PNF22098.1 ADMH02001341 ETN62953.1 ATLV01020674 KE525304 KFB45518.1 KZ288256 PBC30592.1 KK107154 EZA56859.1 NNAY01001402 OXU24099.1 QOIP01000001 RLU26555.1 GL435030 EFN74210.1 KQ976511 KYM82550.1 ADTU01023455 KQ982335 KYQ57564.1 GL888066 EGI68396.1 AXCM01000163 GBRD01001460 GBRD01001459 JAG64361.1 DS235819 EEB17707.1 KQ981280 KYN43485.1 KQ435702 KOX80340.1 GECU01019577 JAS88129.1 KK852463 KDR23323.1 GL449158 EFN83223.1 CH479184 EDW37043.1 CM000361 EDX05092.1 CVRI01000058 CRL02846.1 KQ976885 KYN07498.1 GFXV01000766 MBW12571.1 GL768069 EFZ12431.1 GEZM01047923 JAV76690.1 KU986874 APC26103.1 KU986868 APC26097.1 LJIJ01000095 ODN02817.1 LNIX01000007 OXA52376.1 GDIQ01177444 GDIQ01173250 GDIQ01166409 GDIQ01162156 GDIP01044550 LRGB01000996 JAK78475.1 JAM59165.1 KZS14238.1 GDIQ01027552 JAN67185.1 GDIP01125131 JAL78583.1 GDIP01182023 JAJ41379.1 GDIP01009869 JAM93846.1 GL732530 EFX86255.1 APGK01040537 APGK01040538 APGK01040539 APGK01040540 APGK01040541 APGK01040542 KB740983 ENN76342.1 GBFC01000039 JAC59299.1 GFAC01005516 JAT93672.1 JN974907 AFC88976.1 IAAA01002824 LAA02196.1 GEGO01003217 JAR92187.1
Proteomes
UP000005204
UP000283053
UP000053240
UP000218220
UP000053268
UP000007801
+ More
UP000037069 UP000001070 UP000053825 UP000002282 UP000000803 UP000007798 UP000008711 UP000008792 UP000009192 UP000268350 UP000001819 UP000192221 UP000001292 UP000075840 UP000015103 UP000075882 UP000076408 UP000075885 UP000075881 UP000075886 UP000075884 UP000007062 UP000075900 UP000092461 UP000002358 UP000235965 UP000000673 UP000069272 UP000005203 UP000030765 UP000242457 UP000053097 UP000215335 UP000279307 UP000000311 UP000078540 UP000005205 UP000075809 UP000007755 UP000075920 UP000075883 UP000009046 UP000078541 UP000053105 UP000027135 UP000095301 UP000008237 UP000008744 UP000000304 UP000183832 UP000078542 UP000095300 UP000075903 UP000075902 UP000076407 UP000094527 UP000198287 UP000076858 UP000075880 UP000000305 UP000019118
UP000037069 UP000001070 UP000053825 UP000002282 UP000000803 UP000007798 UP000008711 UP000008792 UP000009192 UP000268350 UP000001819 UP000192221 UP000001292 UP000075840 UP000015103 UP000075882 UP000076408 UP000075885 UP000075881 UP000075886 UP000075884 UP000007062 UP000075900 UP000092461 UP000002358 UP000235965 UP000000673 UP000069272 UP000005203 UP000030765 UP000242457 UP000053097 UP000215335 UP000279307 UP000000311 UP000078540 UP000005205 UP000075809 UP000007755 UP000075920 UP000075883 UP000009046 UP000078541 UP000053105 UP000027135 UP000095301 UP000008237 UP000008744 UP000000304 UP000183832 UP000078542 UP000095300 UP000075903 UP000075902 UP000076407 UP000094527 UP000198287 UP000076858 UP000075880 UP000000305 UP000019118
Interpro
ProteinModelPortal
H9JE90
A0A3S2M9U9
S6DR08
A0A2H1WAC6
A0A194RD58
A0A2A4JSW0
+ More
A0A194PQ25 A0A310SM81 B3MLC7 A0A0L0BVC8 B4JAW8 A0A0L7R583 A0A0R1DST3 A0A0R1DQW3 A0A0R1DSQ2 Q9V3Q9 B4P0X4 B4MZ77 A0A0R1DLG3 B3N5N0 B4LRY4 B4KLC2 A0A0Q5WMT4 W8BH72 A0A3B0KBC3 A0A0R3NQZ1 A0A0Q5WIS6 A0A1W4W038 Q29P68 Q9BML7 A0A0Q5W991 B4HXL4 A0A1S4H3I9 A0A182I0W8 T1I7D6 A0A182KJX6 A0A0J9R2J4 A0A182YN14 A0A182PPU0 A0A182JP69 A0A182Q9X6 A0A182N8X1 V6DNG0 Q7PME5 A0A182RAF4 A0A1B0CTX3 K7IRD3 A0A2J7Q0I1 W5JI42 A0A182F8W2 A0A088ACG8 A0A084W5M4 A0A2A3EH29 A0A026WP30 A0A232F0L8 A0A3L8E2F5 E1ZXC0 A0A195BEF8 A0A158NQP1 A0A151XB41 F4WBW8 A0A182VSS1 A0A182MVL9 A0A0K8TG24 E0VWF6 A0A195FSC0 A0A0M9AA59 A0A1B6IMJ1 A0A067RJX4 A0A1I8MUN8 E2BM97 B4GK77 B4Q5Y9 A0A1J1IUN2 A0A1S4FG88 A0A195D3M2 A0A2H8TER2 A0A1I8PIE9 E9J436 A0A1Y1LWI4 A0A1J0FAU9 A0A182V1I8 A0A182U5X5 A0A182XCP2 A0A1J0FAT2 A0A1D2NC33 A0A226E705 A0A0P5LWC8 A0A0P6GYC0 A0A182J9B7 A0A0P5TKA2 A0A0P5BNQ1 A0A0P6C3F4 E9G2I1 N6UCG3 A0A061QLL2 A0A1E1X357 H9B3X8 A0A2L2Y1Y5 A0A147BN21
A0A194PQ25 A0A310SM81 B3MLC7 A0A0L0BVC8 B4JAW8 A0A0L7R583 A0A0R1DST3 A0A0R1DQW3 A0A0R1DSQ2 Q9V3Q9 B4P0X4 B4MZ77 A0A0R1DLG3 B3N5N0 B4LRY4 B4KLC2 A0A0Q5WMT4 W8BH72 A0A3B0KBC3 A0A0R3NQZ1 A0A0Q5WIS6 A0A1W4W038 Q29P68 Q9BML7 A0A0Q5W991 B4HXL4 A0A1S4H3I9 A0A182I0W8 T1I7D6 A0A182KJX6 A0A0J9R2J4 A0A182YN14 A0A182PPU0 A0A182JP69 A0A182Q9X6 A0A182N8X1 V6DNG0 Q7PME5 A0A182RAF4 A0A1B0CTX3 K7IRD3 A0A2J7Q0I1 W5JI42 A0A182F8W2 A0A088ACG8 A0A084W5M4 A0A2A3EH29 A0A026WP30 A0A232F0L8 A0A3L8E2F5 E1ZXC0 A0A195BEF8 A0A158NQP1 A0A151XB41 F4WBW8 A0A182VSS1 A0A182MVL9 A0A0K8TG24 E0VWF6 A0A195FSC0 A0A0M9AA59 A0A1B6IMJ1 A0A067RJX4 A0A1I8MUN8 E2BM97 B4GK77 B4Q5Y9 A0A1J1IUN2 A0A1S4FG88 A0A195D3M2 A0A2H8TER2 A0A1I8PIE9 E9J436 A0A1Y1LWI4 A0A1J0FAU9 A0A182V1I8 A0A182U5X5 A0A182XCP2 A0A1J0FAT2 A0A1D2NC33 A0A226E705 A0A0P5LWC8 A0A0P6GYC0 A0A182J9B7 A0A0P5TKA2 A0A0P5BNQ1 A0A0P6C3F4 E9G2I1 N6UCG3 A0A061QLL2 A0A1E1X357 H9B3X8 A0A2L2Y1Y5 A0A147BN21
PDB
4MS4
E-value=5.64706e-133,
Score=1218
Ontologies
GO
PANTHER
Topology
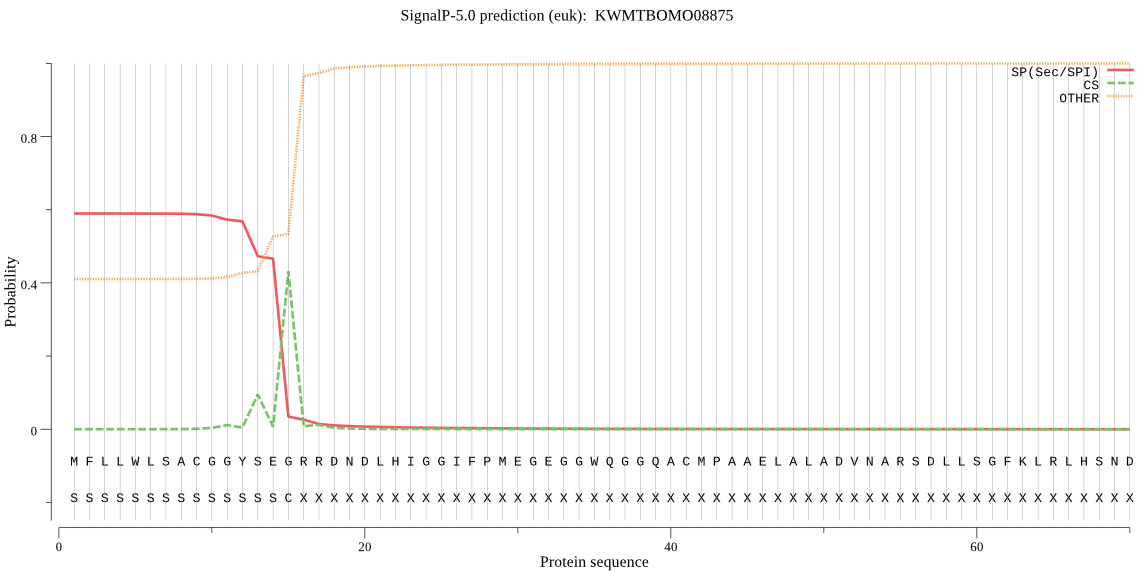
SignalP
Position: 1 - 15,
Likelihood: 0.588624
Length:
784
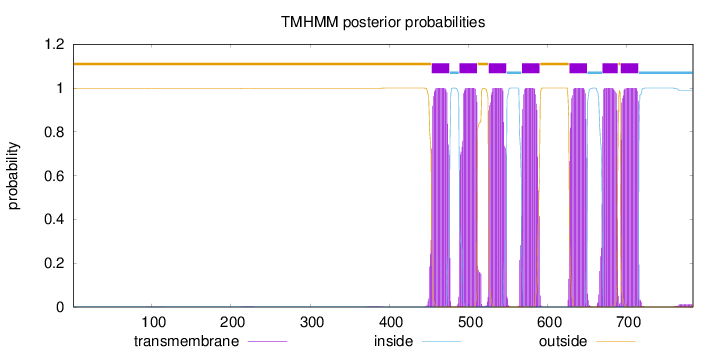
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
154.7048
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00117
outside
1 - 453
TMhelix
454 - 476
inside
477 - 488
TMhelix
489 - 511
outside
512 - 525
TMhelix
526 - 548
inside
549 - 567
TMhelix
568 - 590
outside
591 - 627
TMhelix
628 - 650
inside
651 - 669
TMhelix
670 - 689
outside
690 - 692
TMhelix
693 - 715
inside
716 - 784
Population Genetic Test Statistics
Pi
198.405967
Theta
160.173953
Tajima's D
0.519635
CLR
0.523656
CSRT
0.52002399880006
Interpretation
Uncertain
