Gene
KWMTBOMO08872
Pre Gene Modal
BGIBMGA007839
Annotation
PREDICTED:_1-acyl-sn-glycerol-3-phosphate_acyltransferase_alpha-like_[Amyelois_transitella]
Full name
1-acyl-sn-glycerol-3-phosphate acyltransferase
Location in the cell
Cytoplasmic Reliability : 1.149 PlasmaMembrane Reliability : 1.711
Sequence
CDS
ATGTTCAATATTTGGCATGTTGCCGATAAAGTGGCAGCAGTTGCAAGGAAAGAGATATTTTATGTTTGGCCATTCGGCTTAGCAGCATATTTGGCTGGTGTTGTATTCATAGACCGAAATAATTCTAAAGATGCCTACAAACAGTTGGAAATCACTTCTGAAGTCATGGTGAAGAATAAAACAAAGATCTGGTTGTTCCCTGAAGGCACAAGGAATAAGGATTTTACCAAATTGTTACCATTCAAAAAGGGAGCATTCAACATTGCAGTAGCCGCACAAGTTCCAATTATACCTGTTATATATTCACCGTACTATTTTATCAACAGAAGAAAATGTATTTTTGATAAAGGTCACGTCATAATTCAGTGCCTAGAGCCGGTAACCACCGAAGGTCTGACGATGCGGGACGTGCCCGAGCTGATCGAACGAGTGCGCAACATGATGGACGCCGCATACAAAGAACTTTCGAAAGAGGTACTGAGCGCACTCCCACCTAACTACCCGCTCGCTACGACGGATTGA
Protein
MFNIWHVADKVAAVARKEIFYVWPFGLAAYLAGVVFIDRNNSKDAYKQLEITSEVMVKNKTKIWLFPEGTRNKDFTKLLPFKKGAFNIAVAAQVPIIPVIYSPYYFINRRKCIFDKGHVIIQCLEPVTTEGLTMRDVPELIERVRNMMDAAYKELSKEVLSALPPNYPLATTD
Summary
Catalytic Activity
a 1-acyl-sn-glycero-3-phosphate + an acyl-CoA = a 1,2-diacyl-sn-glycero-3-phosphate + CoA
Similarity
Belongs to the 1-acyl-sn-glycerol-3-phosphate acyltransferase family.
Feature
chain 1-acyl-sn-glycerol-3-phosphate acyltransferase
Uniprot
M4M6X5
H9JE92
A0A3S2P751
A0A291P127
M4M650
A0A2A4IY31
+ More
M4M7W0 A0A194PJM1 A0A194RE42 A0A1Q3G5I5 A0A2A4JVE6 A0A0P0EW45 M4M665 A0A291P186 M4MBK4 A0A291P168 A0A088M9L5 H9JE91 M4M7Y0 A0A212FH93 A0A2H1WCT2 A0A194PKH5 A0A194RDL3 I4DIZ9 A0A3S2LC05 A0A212FH99 A0A2J7QX84 A0A2J7QX82 A0A2H1WJV9 A0A1B6DF96 A0A2A4IWG7 M4M670 A0A067QYR8 A0A1B6J1C5 A0A2P8YIU4 A0A1B6H2W1 A0A1B6KK04 E0VLP3 A0A182VZ27 A0A182T5K3 A0A182YQZ8 A0A182LU07 A0A0L7KTY0 A0A195F6Q8 W5JNG7 A0A151ISA5 A0A0A9WN79 A0A232F3L3 A0A2R7W0J7 A0A182UG95 A0A084VHB1 K7JQK0 A0A182KB72 A0A182XBA1 A0A067R1X9 A0A182LH23 A0A182NCG5 A0A182PRJ4 A0A182I043 Q7PYM8 F4WSQ4 A0A182UYJ7 A0A158NH29 A0A182R6B4 A0A182IJG6 A0A2A3E5E8 B4L214 A0A0L0C157 A0A195AYX3 R4WDV3 A0A0L7L4X5 A0A0L7R5E0 A0A182QQY3 A0A195D1D8 A0A182FNR2 V9IF11 B4JK14 A0A151XDB2 D6X4R5 A0A1B6F296 A0A1W7R8B8 A0A023ERG2 E9GVQ9 A0A087ZWW5 A0A1S4EXH8 A0A1W4V7D8 A0A154PN56 A0A1B6K6S0 E2AA02 A0A026W1G2 Q17MK3 A0A310SG32 B4N1Y9 A0A0R3P544 A0A1L8DYQ1 A0A224XHX0 Q9VYJ4 B4Q1Q5 A0A069DQM8 B3MS13
M4M7W0 A0A194PJM1 A0A194RE42 A0A1Q3G5I5 A0A2A4JVE6 A0A0P0EW45 M4M665 A0A291P186 M4MBK4 A0A291P168 A0A088M9L5 H9JE91 M4M7Y0 A0A212FH93 A0A2H1WCT2 A0A194PKH5 A0A194RDL3 I4DIZ9 A0A3S2LC05 A0A212FH99 A0A2J7QX84 A0A2J7QX82 A0A2H1WJV9 A0A1B6DF96 A0A2A4IWG7 M4M670 A0A067QYR8 A0A1B6J1C5 A0A2P8YIU4 A0A1B6H2W1 A0A1B6KK04 E0VLP3 A0A182VZ27 A0A182T5K3 A0A182YQZ8 A0A182LU07 A0A0L7KTY0 A0A195F6Q8 W5JNG7 A0A151ISA5 A0A0A9WN79 A0A232F3L3 A0A2R7W0J7 A0A182UG95 A0A084VHB1 K7JQK0 A0A182KB72 A0A182XBA1 A0A067R1X9 A0A182LH23 A0A182NCG5 A0A182PRJ4 A0A182I043 Q7PYM8 F4WSQ4 A0A182UYJ7 A0A158NH29 A0A182R6B4 A0A182IJG6 A0A2A3E5E8 B4L214 A0A0L0C157 A0A195AYX3 R4WDV3 A0A0L7L4X5 A0A0L7R5E0 A0A182QQY3 A0A195D1D8 A0A182FNR2 V9IF11 B4JK14 A0A151XDB2 D6X4R5 A0A1B6F296 A0A1W7R8B8 A0A023ERG2 E9GVQ9 A0A087ZWW5 A0A1S4EXH8 A0A1W4V7D8 A0A154PN56 A0A1B6K6S0 E2AA02 A0A026W1G2 Q17MK3 A0A310SG32 B4N1Y9 A0A0R3P544 A0A1L8DYQ1 A0A224XHX0 Q9VYJ4 B4Q1Q5 A0A069DQM8 B3MS13
EC Number
2.3.1.51
Pubmed
23294019
19121390
28986331
26354079
26445454
26385554
+ More
22118469 22651552 24845553 29403074 20566863 25244985 26227816 20920257 23761445 25401762 26823975 28648823 24438588 20075255 20966253 12364791 14747013 17210077 21719571 21347285 17994087 18057021 26108605 23691247 18362917 19820115 24945155 21292972 20798317 24508170 30249741 17510324 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 26334808
22118469 22651552 24845553 29403074 20566863 25244985 26227816 20920257 23761445 25401762 26823975 28648823 24438588 20075255 20966253 12364791 14747013 17210077 21719571 21347285 17994087 18057021 26108605 23691247 18362917 19820115 24945155 21292972 20798317 24508170 30249741 17510324 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 26334808
EMBL
JX847007
AGG55024.1
BABH01002768
RSAL01000255
RVE43422.1
MF687666
+ More
ATJ44592.1 JX846994 AGG55011.1 NWSH01005665 PCG64070.1 JX846978 AGG54995.1 KQ459602 KPI93233.1 KQ460367 KPJ15550.1 GEYN01000176 JAV44813.1 NWSH01000503 PCG75995.1 KT261723 ALJ30263.1 JX846980 AGG54997.1 MF706182 ATJ44609.1 JX846996 AGG55013.1 MF687664 ATJ44590.1 KJ579238 AIN34714.1 BABH01002767 JX847008 AGG55025.1 AGBW02008520 OWR53099.1 ODYU01007807 SOQ50885.1 KPI93234.1 KPJ15549.1 AK401267 BAM17889.1 RVE43421.1 OWR53100.1 NEVH01009384 PNF33195.1 PNF33197.1 ODYU01009125 SOQ53328.1 GEDC01012930 JAS24368.1 PCG64071.1 JX846995 AGG55012.1 KK852869 KDR14660.1 GECU01014744 JAS92962.1 PYGN01000565 PSN44158.1 GECZ01000809 JAS68960.1 GEBQ01028190 JAT11787.1 DS235277 EEB14299.1 AXCM01005544 JTDY01005891 KOB66506.1 KQ981768 KYN35877.1 ADMH02001019 ETN64415.1 KQ981091 KYN09551.1 GBHO01033684 GBHO01033683 GBHO01033682 GBRD01009564 GBRD01009563 GBRD01009562 GDHC01008152 JAG09920.1 JAG09921.1 JAG09922.1 JAG56260.1 JAQ10477.1 NNAY01001120 OXU25070.1 KK854198 PTY12968.1 ATLV01013150 KE524842 KFB37355.1 KK852771 KDR16862.1 APCN01002042 AAAB01008987 EAA01231.5 GL888327 EGI62764.1 ADTU01015495 KZ288388 PBC26399.1 CH933810 EDW07735.1 JRES01001143 KNC25189.1 KQ976698 KYM77443.1 AK417993 BAN21208.1 JTDY01003006 KOB70324.1 KQ414654 KOC65976.1 AXCN02000016 KQ976973 KYN06682.1 JR040716 JR040717 AEY59257.1 AEY59258.1 CH916370 EDV99916.1 KQ982294 KYQ58362.1 KQ971380 EEZ97656.1 GECZ01025444 JAS44325.1 GEHC01000268 JAV47377.1 GAPW01002344 JAC11254.1 GL732568 EFX76482.1 KQ434998 KZC13289.1 GECU01034469 GECU01000612 JAS73237.1 JAT07095.1 GL437990 EFN69731.1 KK107566 QOIP01000006 EZA48899.1 RLU21916.1 CH477205 EAT47921.1 KQ760357 OAD60753.1 CH963925 EDW78378.2 CH379064 KRT06685.1 GFDF01002662 JAV11422.1 GFTR01004341 JAW12085.1 AE014298 AY118588 AAF48201.1 AAM49957.1 AAN09316.1 CM000162 EDX01496.1 GBGD01002541 JAC86348.1 CH902622 EDV34568.1 KPU75201.1
ATJ44592.1 JX846994 AGG55011.1 NWSH01005665 PCG64070.1 JX846978 AGG54995.1 KQ459602 KPI93233.1 KQ460367 KPJ15550.1 GEYN01000176 JAV44813.1 NWSH01000503 PCG75995.1 KT261723 ALJ30263.1 JX846980 AGG54997.1 MF706182 ATJ44609.1 JX846996 AGG55013.1 MF687664 ATJ44590.1 KJ579238 AIN34714.1 BABH01002767 JX847008 AGG55025.1 AGBW02008520 OWR53099.1 ODYU01007807 SOQ50885.1 KPI93234.1 KPJ15549.1 AK401267 BAM17889.1 RVE43421.1 OWR53100.1 NEVH01009384 PNF33195.1 PNF33197.1 ODYU01009125 SOQ53328.1 GEDC01012930 JAS24368.1 PCG64071.1 JX846995 AGG55012.1 KK852869 KDR14660.1 GECU01014744 JAS92962.1 PYGN01000565 PSN44158.1 GECZ01000809 JAS68960.1 GEBQ01028190 JAT11787.1 DS235277 EEB14299.1 AXCM01005544 JTDY01005891 KOB66506.1 KQ981768 KYN35877.1 ADMH02001019 ETN64415.1 KQ981091 KYN09551.1 GBHO01033684 GBHO01033683 GBHO01033682 GBRD01009564 GBRD01009563 GBRD01009562 GDHC01008152 JAG09920.1 JAG09921.1 JAG09922.1 JAG56260.1 JAQ10477.1 NNAY01001120 OXU25070.1 KK854198 PTY12968.1 ATLV01013150 KE524842 KFB37355.1 KK852771 KDR16862.1 APCN01002042 AAAB01008987 EAA01231.5 GL888327 EGI62764.1 ADTU01015495 KZ288388 PBC26399.1 CH933810 EDW07735.1 JRES01001143 KNC25189.1 KQ976698 KYM77443.1 AK417993 BAN21208.1 JTDY01003006 KOB70324.1 KQ414654 KOC65976.1 AXCN02000016 KQ976973 KYN06682.1 JR040716 JR040717 AEY59257.1 AEY59258.1 CH916370 EDV99916.1 KQ982294 KYQ58362.1 KQ971380 EEZ97656.1 GECZ01025444 JAS44325.1 GEHC01000268 JAV47377.1 GAPW01002344 JAC11254.1 GL732568 EFX76482.1 KQ434998 KZC13289.1 GECU01034469 GECU01000612 JAS73237.1 JAT07095.1 GL437990 EFN69731.1 KK107566 QOIP01000006 EZA48899.1 RLU21916.1 CH477205 EAT47921.1 KQ760357 OAD60753.1 CH963925 EDW78378.2 CH379064 KRT06685.1 GFDF01002662 JAV11422.1 GFTR01004341 JAW12085.1 AE014298 AY118588 AAF48201.1 AAM49957.1 AAN09316.1 CM000162 EDX01496.1 GBGD01002541 JAC86348.1 CH902622 EDV34568.1 KPU75201.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000053240
UP000007151
+ More
UP000235965 UP000027135 UP000245037 UP000009046 UP000075920 UP000075901 UP000076408 UP000075883 UP000037510 UP000078541 UP000000673 UP000078492 UP000215335 UP000075902 UP000030765 UP000002358 UP000075881 UP000076407 UP000075882 UP000075884 UP000075885 UP000075840 UP000007062 UP000007755 UP000075903 UP000005205 UP000075900 UP000075880 UP000242457 UP000009192 UP000037069 UP000078540 UP000053825 UP000075886 UP000078542 UP000069272 UP000001070 UP000075809 UP000007266 UP000000305 UP000005203 UP000192221 UP000076502 UP000000311 UP000053097 UP000279307 UP000008820 UP000007798 UP000001819 UP000000803 UP000002282 UP000007801
UP000235965 UP000027135 UP000245037 UP000009046 UP000075920 UP000075901 UP000076408 UP000075883 UP000037510 UP000078541 UP000000673 UP000078492 UP000215335 UP000075902 UP000030765 UP000002358 UP000075881 UP000076407 UP000075882 UP000075884 UP000075885 UP000075840 UP000007062 UP000007755 UP000075903 UP000005205 UP000075900 UP000075880 UP000242457 UP000009192 UP000037069 UP000078540 UP000053825 UP000075886 UP000078542 UP000069272 UP000001070 UP000075809 UP000007266 UP000000305 UP000005203 UP000192221 UP000076502 UP000000311 UP000053097 UP000279307 UP000008820 UP000007798 UP000001819 UP000000803 UP000002282 UP000007801
Pfam
PF01553 Acyltransferase
ProteinModelPortal
M4M6X5
H9JE92
A0A3S2P751
A0A291P127
M4M650
A0A2A4IY31
+ More
M4M7W0 A0A194PJM1 A0A194RE42 A0A1Q3G5I5 A0A2A4JVE6 A0A0P0EW45 M4M665 A0A291P186 M4MBK4 A0A291P168 A0A088M9L5 H9JE91 M4M7Y0 A0A212FH93 A0A2H1WCT2 A0A194PKH5 A0A194RDL3 I4DIZ9 A0A3S2LC05 A0A212FH99 A0A2J7QX84 A0A2J7QX82 A0A2H1WJV9 A0A1B6DF96 A0A2A4IWG7 M4M670 A0A067QYR8 A0A1B6J1C5 A0A2P8YIU4 A0A1B6H2W1 A0A1B6KK04 E0VLP3 A0A182VZ27 A0A182T5K3 A0A182YQZ8 A0A182LU07 A0A0L7KTY0 A0A195F6Q8 W5JNG7 A0A151ISA5 A0A0A9WN79 A0A232F3L3 A0A2R7W0J7 A0A182UG95 A0A084VHB1 K7JQK0 A0A182KB72 A0A182XBA1 A0A067R1X9 A0A182LH23 A0A182NCG5 A0A182PRJ4 A0A182I043 Q7PYM8 F4WSQ4 A0A182UYJ7 A0A158NH29 A0A182R6B4 A0A182IJG6 A0A2A3E5E8 B4L214 A0A0L0C157 A0A195AYX3 R4WDV3 A0A0L7L4X5 A0A0L7R5E0 A0A182QQY3 A0A195D1D8 A0A182FNR2 V9IF11 B4JK14 A0A151XDB2 D6X4R5 A0A1B6F296 A0A1W7R8B8 A0A023ERG2 E9GVQ9 A0A087ZWW5 A0A1S4EXH8 A0A1W4V7D8 A0A154PN56 A0A1B6K6S0 E2AA02 A0A026W1G2 Q17MK3 A0A310SG32 B4N1Y9 A0A0R3P544 A0A1L8DYQ1 A0A224XHX0 Q9VYJ4 B4Q1Q5 A0A069DQM8 B3MS13
M4M7W0 A0A194PJM1 A0A194RE42 A0A1Q3G5I5 A0A2A4JVE6 A0A0P0EW45 M4M665 A0A291P186 M4MBK4 A0A291P168 A0A088M9L5 H9JE91 M4M7Y0 A0A212FH93 A0A2H1WCT2 A0A194PKH5 A0A194RDL3 I4DIZ9 A0A3S2LC05 A0A212FH99 A0A2J7QX84 A0A2J7QX82 A0A2H1WJV9 A0A1B6DF96 A0A2A4IWG7 M4M670 A0A067QYR8 A0A1B6J1C5 A0A2P8YIU4 A0A1B6H2W1 A0A1B6KK04 E0VLP3 A0A182VZ27 A0A182T5K3 A0A182YQZ8 A0A182LU07 A0A0L7KTY0 A0A195F6Q8 W5JNG7 A0A151ISA5 A0A0A9WN79 A0A232F3L3 A0A2R7W0J7 A0A182UG95 A0A084VHB1 K7JQK0 A0A182KB72 A0A182XBA1 A0A067R1X9 A0A182LH23 A0A182NCG5 A0A182PRJ4 A0A182I043 Q7PYM8 F4WSQ4 A0A182UYJ7 A0A158NH29 A0A182R6B4 A0A182IJG6 A0A2A3E5E8 B4L214 A0A0L0C157 A0A195AYX3 R4WDV3 A0A0L7L4X5 A0A0L7R5E0 A0A182QQY3 A0A195D1D8 A0A182FNR2 V9IF11 B4JK14 A0A151XDB2 D6X4R5 A0A1B6F296 A0A1W7R8B8 A0A023ERG2 E9GVQ9 A0A087ZWW5 A0A1S4EXH8 A0A1W4V7D8 A0A154PN56 A0A1B6K6S0 E2AA02 A0A026W1G2 Q17MK3 A0A310SG32 B4N1Y9 A0A0R3P544 A0A1L8DYQ1 A0A224XHX0 Q9VYJ4 B4Q1Q5 A0A069DQM8 B3MS13
PDB
5KYM
E-value=4.24754e-11,
Score=159
Ontologies
PATHWAY
GO
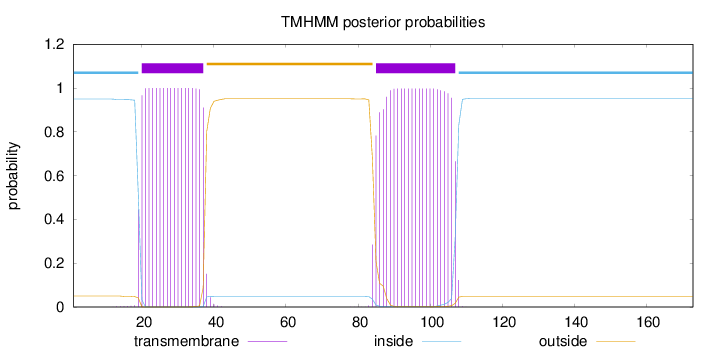
Topology
Length:
173
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
41.03113
Exp number, first 60 AAs:
18.55083
Total prob of N-in:
0.95029
POSSIBLE N-term signal
sequence
inside
1 - 19
TMhelix
20 - 37
outside
38 - 84
TMhelix
85 - 107
inside
108 - 173
Population Genetic Test Statistics
Pi
178.285068
Theta
164.394939
Tajima's D
-0.452278
CLR
1.801653
CSRT
0.248137593120344
Interpretation
Uncertain
