Pre Gene Modal
BGIBMGA007706
Annotation
PREDICTED:_leucine-rich_repeat-containing_protein_57_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 2.175
Sequence
CDS
ATGGGGAACGCTGGTTTAAAACAACATTATGAAACTGCATCGAAAACTGGTGTATTGCAAATAGCTGATTGTAAGCTTAAAGAAATTCCCATCGACGTATTCAGTTTATGTGATGTGCTACGAAATCTAGATCTGTCAAAAAACAAACTTTCTGTTTTACCTGACGATTTAAGCAAATTAAAACACCTGAAACAATTAAATTTAGATAGTAATAAACTTCAAGCTTTACCTGAATCACTCATAAACTTAAAAAAACTTGAAGTTTTAAATATATCAAACAACTTTGTACAAACACTCCCAGTTTGTTTGAAGAATTTAAATAATTTAAAACAGGTTTATGCAAGCAATAATAAATTACAGGCTTTTCCGAAACAAATACTTGGATTACAGAGCTTAGATGTTTTAGATCTGTCACATAATAAGATTACAGAGATTCCATATGGAATGTCAGAATTATATGTGATAGAGTTAAATCTGAGTCAAAATGAAATATCTTTGATTGGAGAAGACTTACACCAAGCTCCAAGATTAAAGATATTGCGATTGGATGAGAACTGTTTGAGTTTGGATGAAATCAAACCAAGTTTGCTTCGAGATTCCAAGATACACACTGTTAGTGTAGATGGTAATTTATTTGAGCCTAAACAGTTAGCATCACTTGAGGGCTATAATGAATACAGTGAAAGATATACTGCAATGAAGAAAAAAATGTTTTGA
Protein
MGNAGLKQHYETASKTGVLQIADCKLKEIPIDVFSLCDVLRNLDLSKNKLSVLPDDLSKLKHLKQLNLDSNKLQALPESLINLKKLEVLNISNNFVQTLPVCLKNLNNLKQVYASNNKLQAFPKQILGLQSLDVLDLSHNKITEIPYGMSELYVIELNLSQNEISLIGEDLHQAPRLKILRLDENCLSLDEIKPSLLRDSKIHTVSVDGNLFEPKQLASLEGYNEYSERYTAMKKKMF
Summary
Uniprot
H9JDV9
A0A2H1WH15
A0A212FH83
A0A0L7L0P3
A0A437AZX0
A0A194PII8
+ More
A0A194RD64 A0A067QWT0 A0A2J7PJ32 A0A0L7QYZ0 A0A2P8YLN6 A0A026WF94 A0A0M8ZQL3 U5EL17 A0A232ET50 A0A0C9Q1M4 K7IZN7 A0A088ACM2 A0A2A3EP03 A0A310SH01 A0A154PJJ1 A0A195ERM0 A0A1J1I075 E2AFH1 A0A0J7KYZ3 E2C5V2 A0A195BR21 A0A195CND2 A0A1W4WT49 A0A195DHT4 F4WKD2 A0A0A1WQC5 W8B810 A0A158P1Q2 A0A0K8WDP5 A0A034V8F9 A0A1Y1K4U9 A0A151XAS9 E9J3Y3 A0A1B6EB31 A0A1L8DB69 A0A034V677 A0A0V0GC11 B4JLB7 A0A0K8UM95 A0A1W4UWD2 B3NXX9 B4NCS2 A0A0L0BWI8 A0A1L8DCR9 B4Q1D1 A0A084W2Z9 A0A182M5W2 B4R5W0 B4IHI2 Q9W3T9 A0A182PGZ5 A0A0P4ZMD2 A0A182JZM5 J3JYE1 A0A182VWE1 B0X7R9 A0A1A9XRV7 A0A1B0AP68 A0A023EKR0 A0A0P5KIT9 A0A1A9ZE58 A0A182UIF8 A0A182QMZ7 A0A182HL12 A0A182R505 A0A182V2A0 Q7PXN5 A0A182WS64 A0A336ML58 B4M6N2 A0A182L9D1 A0A093BDG7 E0VWT8 B4L204 A0A1A9UUN7 A0A093GAJ3 L7MDG9 A0A1E1XRJ6 B3MS52 E9HRR4 A0A091J250 A0A182YB91 A0A0K8TLW7 A0A0Q3RJI6 A0A1A9WPJ4 A0A093K7D1 A0A093FQX2 A0A182FRR5 A0A2M4BY96 A0A2M4AY58 A0A293LCF0 A0A2M3ZJA3 A0A2R5LGS1
A0A194RD64 A0A067QWT0 A0A2J7PJ32 A0A0L7QYZ0 A0A2P8YLN6 A0A026WF94 A0A0M8ZQL3 U5EL17 A0A232ET50 A0A0C9Q1M4 K7IZN7 A0A088ACM2 A0A2A3EP03 A0A310SH01 A0A154PJJ1 A0A195ERM0 A0A1J1I075 E2AFH1 A0A0J7KYZ3 E2C5V2 A0A195BR21 A0A195CND2 A0A1W4WT49 A0A195DHT4 F4WKD2 A0A0A1WQC5 W8B810 A0A158P1Q2 A0A0K8WDP5 A0A034V8F9 A0A1Y1K4U9 A0A151XAS9 E9J3Y3 A0A1B6EB31 A0A1L8DB69 A0A034V677 A0A0V0GC11 B4JLB7 A0A0K8UM95 A0A1W4UWD2 B3NXX9 B4NCS2 A0A0L0BWI8 A0A1L8DCR9 B4Q1D1 A0A084W2Z9 A0A182M5W2 B4R5W0 B4IHI2 Q9W3T9 A0A182PGZ5 A0A0P4ZMD2 A0A182JZM5 J3JYE1 A0A182VWE1 B0X7R9 A0A1A9XRV7 A0A1B0AP68 A0A023EKR0 A0A0P5KIT9 A0A1A9ZE58 A0A182UIF8 A0A182QMZ7 A0A182HL12 A0A182R505 A0A182V2A0 Q7PXN5 A0A182WS64 A0A336ML58 B4M6N2 A0A182L9D1 A0A093BDG7 E0VWT8 B4L204 A0A1A9UUN7 A0A093GAJ3 L7MDG9 A0A1E1XRJ6 B3MS52 E9HRR4 A0A091J250 A0A182YB91 A0A0K8TLW7 A0A0Q3RJI6 A0A1A9WPJ4 A0A093K7D1 A0A093FQX2 A0A182FRR5 A0A2M4BY96 A0A2M4AY58 A0A293LCF0 A0A2M3ZJA3 A0A2R5LGS1
Pubmed
19121390
22118469
26227816
26354079
24845553
29403074
+ More
24508170 30249741 28648823 20075255 20798317 21719571 25830018 24495485 21347285 25348373 28004739 21282665 17994087 26108605 17550304 24438588 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22516182 24945155 26483478 12364791 14747013 17210077 20966253 20566863 25576852 29209593 21292972 25244985 26369729
24508170 30249741 28648823 20075255 20798317 21719571 25830018 24495485 21347285 25348373 28004739 21282665 17994087 26108605 17550304 24438588 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22516182 24945155 26483478 12364791 14747013 17210077 20966253 20566863 25576852 29209593 21292972 25244985 26369729
EMBL
BABH01002773
ODYU01008515
SOQ52182.1
AGBW02008520
OWR53102.1
JTDY01003880
+ More
KOB68881.1 RSAL01000255 RVE43425.1 KQ459602 KPI93231.1 KQ460367 KPJ15552.1 KK853274 KDR09114.1 NEVH01024952 PNF16329.1 KQ414685 KOC63781.1 PYGN01000508 PSN45158.1 KK107274 QOIP01000008 EZA53684.1 RLU19216.1 KQ436003 KOX67633.1 GANO01001617 JAB58254.1 NNAY01002329 OXU21538.1 GBYB01007798 JAG77565.1 KZ288204 PBC33244.1 KQ766904 OAD53545.1 KQ434936 KZC11983.1 KQ982021 KYN30524.1 CVRI01000024 CRK92214.1 GL439108 EFN67864.1 LBMM01001944 KMQ95499.1 GL452837 EFN76680.1 KQ976419 KYM89333.1 KQ977595 KYN01594.1 KQ980824 KYN12463.1 GL888199 EGI65356.1 GBXI01013659 JAD00633.1 GAMC01009200 GAMC01009198 JAB97355.1 ADTU01006747 GDHF01003334 JAI48980.1 GAKP01020158 JAC38794.1 GEZM01092697 JAV56492.1 KQ982335 KYQ57483.1 GL768069 EFZ12440.1 GEDC01002167 JAS35131.1 GFDF01010373 JAV03711.1 GAKP01020156 JAC38796.1 GECL01000581 JAP05543.1 CH916370 EDW00370.1 GDHF01024517 GDHF01000534 JAI27797.1 JAI51780.1 CH954180 EDV47430.1 CH964239 EDW82631.1 JRES01001325 KNC23629.1 GFDF01009832 JAV04252.1 CM000162 EDX02420.1 ATLV01019742 KE525279 KFB44593.1 AXCM01003927 CM000366 EDX17296.1 CH480839 EDW49333.1 AE014298 AY060823 AAF46224.2 AAL28371.1 GDIP01214492 LRGB01000115 JAJ08910.1 KZS20812.1 BT128267 AEE63227.1 DS232459 EDS42081.1 JXJN01001236 JXUM01022600 GAPW01003851 KQ560636 JAC09747.1 KXJ81492.1 GDIQ01190582 JAK61143.1 AXCN02001524 APCN01000878 AAAB01008987 EAA00886.2 UFQT01000718 SSX26748.1 CH940653 EDW62449.2 KN125638 KFU83536.1 DS235824 EEB17844.1 CH933810 EDW07725.2 KL215719 KFV66193.1 GACK01003014 JAA62020.1 GFAA01001758 JAU01677.1 CH902622 EDV34607.1 GL732740 EFX65577.1 KL218481 KFP05866.1 GDAI01002254 JAI15349.1 LMAW01000714 KQK85865.1 KL206840 KFV86329.1 KK399431 KFV59085.1 GGFJ01008914 MBW58055.1 GGFK01012403 MBW45724.1 GFWV01006367 MAA31097.1 GGFM01007906 MBW28657.1 GGLE01004532 MBY08658.1
KOB68881.1 RSAL01000255 RVE43425.1 KQ459602 KPI93231.1 KQ460367 KPJ15552.1 KK853274 KDR09114.1 NEVH01024952 PNF16329.1 KQ414685 KOC63781.1 PYGN01000508 PSN45158.1 KK107274 QOIP01000008 EZA53684.1 RLU19216.1 KQ436003 KOX67633.1 GANO01001617 JAB58254.1 NNAY01002329 OXU21538.1 GBYB01007798 JAG77565.1 KZ288204 PBC33244.1 KQ766904 OAD53545.1 KQ434936 KZC11983.1 KQ982021 KYN30524.1 CVRI01000024 CRK92214.1 GL439108 EFN67864.1 LBMM01001944 KMQ95499.1 GL452837 EFN76680.1 KQ976419 KYM89333.1 KQ977595 KYN01594.1 KQ980824 KYN12463.1 GL888199 EGI65356.1 GBXI01013659 JAD00633.1 GAMC01009200 GAMC01009198 JAB97355.1 ADTU01006747 GDHF01003334 JAI48980.1 GAKP01020158 JAC38794.1 GEZM01092697 JAV56492.1 KQ982335 KYQ57483.1 GL768069 EFZ12440.1 GEDC01002167 JAS35131.1 GFDF01010373 JAV03711.1 GAKP01020156 JAC38796.1 GECL01000581 JAP05543.1 CH916370 EDW00370.1 GDHF01024517 GDHF01000534 JAI27797.1 JAI51780.1 CH954180 EDV47430.1 CH964239 EDW82631.1 JRES01001325 KNC23629.1 GFDF01009832 JAV04252.1 CM000162 EDX02420.1 ATLV01019742 KE525279 KFB44593.1 AXCM01003927 CM000366 EDX17296.1 CH480839 EDW49333.1 AE014298 AY060823 AAF46224.2 AAL28371.1 GDIP01214492 LRGB01000115 JAJ08910.1 KZS20812.1 BT128267 AEE63227.1 DS232459 EDS42081.1 JXJN01001236 JXUM01022600 GAPW01003851 KQ560636 JAC09747.1 KXJ81492.1 GDIQ01190582 JAK61143.1 AXCN02001524 APCN01000878 AAAB01008987 EAA00886.2 UFQT01000718 SSX26748.1 CH940653 EDW62449.2 KN125638 KFU83536.1 DS235824 EEB17844.1 CH933810 EDW07725.2 KL215719 KFV66193.1 GACK01003014 JAA62020.1 GFAA01001758 JAU01677.1 CH902622 EDV34607.1 GL732740 EFX65577.1 KL218481 KFP05866.1 GDAI01002254 JAI15349.1 LMAW01000714 KQK85865.1 KL206840 KFV86329.1 KK399431 KFV59085.1 GGFJ01008914 MBW58055.1 GGFK01012403 MBW45724.1 GFWV01006367 MAA31097.1 GGFM01007906 MBW28657.1 GGLE01004532 MBY08658.1
Proteomes
UP000005204
UP000007151
UP000037510
UP000283053
UP000053268
UP000053240
+ More
UP000027135 UP000235965 UP000053825 UP000245037 UP000053097 UP000279307 UP000053105 UP000215335 UP000002358 UP000005203 UP000242457 UP000076502 UP000078541 UP000183832 UP000000311 UP000036403 UP000008237 UP000078540 UP000078542 UP000192223 UP000078492 UP000007755 UP000005205 UP000075809 UP000001070 UP000192221 UP000008711 UP000007798 UP000037069 UP000002282 UP000030765 UP000075883 UP000000304 UP000001292 UP000000803 UP000075885 UP000076858 UP000075881 UP000075920 UP000002320 UP000092443 UP000092460 UP000069940 UP000249989 UP000092445 UP000075902 UP000075886 UP000075840 UP000075900 UP000075903 UP000007062 UP000076407 UP000008792 UP000075882 UP000009046 UP000009192 UP000078200 UP000053875 UP000007801 UP000000305 UP000054308 UP000076408 UP000051836 UP000091820 UP000053584 UP000069272
UP000027135 UP000235965 UP000053825 UP000245037 UP000053097 UP000279307 UP000053105 UP000215335 UP000002358 UP000005203 UP000242457 UP000076502 UP000078541 UP000183832 UP000000311 UP000036403 UP000008237 UP000078540 UP000078542 UP000192223 UP000078492 UP000007755 UP000005205 UP000075809 UP000001070 UP000192221 UP000008711 UP000007798 UP000037069 UP000002282 UP000030765 UP000075883 UP000000304 UP000001292 UP000000803 UP000075885 UP000076858 UP000075881 UP000075920 UP000002320 UP000092443 UP000092460 UP000069940 UP000249989 UP000092445 UP000075902 UP000075886 UP000075840 UP000075900 UP000075903 UP000007062 UP000076407 UP000008792 UP000075882 UP000009046 UP000009192 UP000078200 UP000053875 UP000007801 UP000000305 UP000054308 UP000076408 UP000051836 UP000091820 UP000053584 UP000069272
Interpro
Gene 3D
ProteinModelPortal
H9JDV9
A0A2H1WH15
A0A212FH83
A0A0L7L0P3
A0A437AZX0
A0A194PII8
+ More
A0A194RD64 A0A067QWT0 A0A2J7PJ32 A0A0L7QYZ0 A0A2P8YLN6 A0A026WF94 A0A0M8ZQL3 U5EL17 A0A232ET50 A0A0C9Q1M4 K7IZN7 A0A088ACM2 A0A2A3EP03 A0A310SH01 A0A154PJJ1 A0A195ERM0 A0A1J1I075 E2AFH1 A0A0J7KYZ3 E2C5V2 A0A195BR21 A0A195CND2 A0A1W4WT49 A0A195DHT4 F4WKD2 A0A0A1WQC5 W8B810 A0A158P1Q2 A0A0K8WDP5 A0A034V8F9 A0A1Y1K4U9 A0A151XAS9 E9J3Y3 A0A1B6EB31 A0A1L8DB69 A0A034V677 A0A0V0GC11 B4JLB7 A0A0K8UM95 A0A1W4UWD2 B3NXX9 B4NCS2 A0A0L0BWI8 A0A1L8DCR9 B4Q1D1 A0A084W2Z9 A0A182M5W2 B4R5W0 B4IHI2 Q9W3T9 A0A182PGZ5 A0A0P4ZMD2 A0A182JZM5 J3JYE1 A0A182VWE1 B0X7R9 A0A1A9XRV7 A0A1B0AP68 A0A023EKR0 A0A0P5KIT9 A0A1A9ZE58 A0A182UIF8 A0A182QMZ7 A0A182HL12 A0A182R505 A0A182V2A0 Q7PXN5 A0A182WS64 A0A336ML58 B4M6N2 A0A182L9D1 A0A093BDG7 E0VWT8 B4L204 A0A1A9UUN7 A0A093GAJ3 L7MDG9 A0A1E1XRJ6 B3MS52 E9HRR4 A0A091J250 A0A182YB91 A0A0K8TLW7 A0A0Q3RJI6 A0A1A9WPJ4 A0A093K7D1 A0A093FQX2 A0A182FRR5 A0A2M4BY96 A0A2M4AY58 A0A293LCF0 A0A2M3ZJA3 A0A2R5LGS1
A0A194RD64 A0A067QWT0 A0A2J7PJ32 A0A0L7QYZ0 A0A2P8YLN6 A0A026WF94 A0A0M8ZQL3 U5EL17 A0A232ET50 A0A0C9Q1M4 K7IZN7 A0A088ACM2 A0A2A3EP03 A0A310SH01 A0A154PJJ1 A0A195ERM0 A0A1J1I075 E2AFH1 A0A0J7KYZ3 E2C5V2 A0A195BR21 A0A195CND2 A0A1W4WT49 A0A195DHT4 F4WKD2 A0A0A1WQC5 W8B810 A0A158P1Q2 A0A0K8WDP5 A0A034V8F9 A0A1Y1K4U9 A0A151XAS9 E9J3Y3 A0A1B6EB31 A0A1L8DB69 A0A034V677 A0A0V0GC11 B4JLB7 A0A0K8UM95 A0A1W4UWD2 B3NXX9 B4NCS2 A0A0L0BWI8 A0A1L8DCR9 B4Q1D1 A0A084W2Z9 A0A182M5W2 B4R5W0 B4IHI2 Q9W3T9 A0A182PGZ5 A0A0P4ZMD2 A0A182JZM5 J3JYE1 A0A182VWE1 B0X7R9 A0A1A9XRV7 A0A1B0AP68 A0A023EKR0 A0A0P5KIT9 A0A1A9ZE58 A0A182UIF8 A0A182QMZ7 A0A182HL12 A0A182R505 A0A182V2A0 Q7PXN5 A0A182WS64 A0A336ML58 B4M6N2 A0A182L9D1 A0A093BDG7 E0VWT8 B4L204 A0A1A9UUN7 A0A093GAJ3 L7MDG9 A0A1E1XRJ6 B3MS52 E9HRR4 A0A091J250 A0A182YB91 A0A0K8TLW7 A0A0Q3RJI6 A0A1A9WPJ4 A0A093K7D1 A0A093FQX2 A0A182FRR5 A0A2M4BY96 A0A2M4AY58 A0A293LCF0 A0A2M3ZJA3 A0A2R5LGS1
PDB
4U06
E-value=2.75562e-16,
Score=206
Ontologies
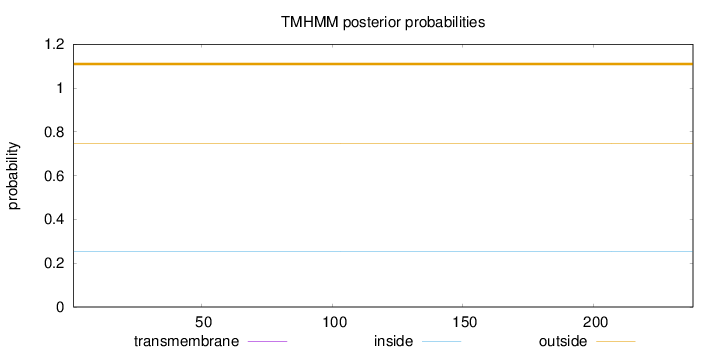
Topology
Length:
238
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00059
Exp number, first 60 AAs:
0
Total prob of N-in:
0.25445
outside
1 - 238
Population Genetic Test Statistics
Pi
258.66944
Theta
201.966015
Tajima's D
0.883764
CLR
0
CSRT
0.627918604069796
Interpretation
Possibly Positive selection
