Gene
KWMTBOMO08867
Pre Gene Modal
BGIBMGA007841
Annotation
high-affinity_copper_uptake_protein_1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.395
Sequence
CDS
ATGGACGAACATCATGAACACCACAATATGGATATGGGCCATGCTGGGCATGAACACCATGGGATTGATCATTCAAGACATATAGGCCACCAAATGCCTATTAATGGACTTCTTAATTCATCATATAACAGAATTGTGCACAACACAGATATGAATGATCACAATGTCCACACTTTTTCAGGCCATGGTGATCACAGTAGCCACAATATGGGCATGTCCATGACGTTTCACGGTGGCTACATTGAAACTATTCTCTTCTCTTGGTGGAACGTAACAGAAGTTGGTGAATTTGTGGGATCATTTTTTGCAATATTTATTATAGCTTTGCTCTATGAAGGCCTTAAATATTATAGAAAGCATTTACTATGGAAGACATATGCAGGACTACAGTATTGTGCCGTAGCACCACCAGACAAGGGAGTTGCCAATATTTGTGCAGCAGATGAACCACAAATTGTACAGCCAATTCCTCATATGCTTGAAAGAAATGTTCCTACGATGATGAGCACAGCACACGCATGGCAAACGATACTTCATGGGGTTCAAGTGCTAGTTAGTTATATGTTGATGTTGGTTTTTATGACTTACAACACATGGCTGTGCGCTGCTGTAGTATTAGGATCAGCGACAGGGTACTTCCTCTTTGGTTGGCGCGAATCGGTAGTTGTTGACTTCACTGAACATTGTCACTGA
Protein
MDEHHEHHNMDMGHAGHEHHGIDHSRHIGHQMPINGLLNSSYNRIVHNTDMNDHNVHTFSGHGDHSSHNMGMSMTFHGGYIETILFSWWNVTEVGEFVGSFFAIFIIALLYEGLKYYRKHLLWKTYAGLQYCAVAPPDKGVANICAADEPQIVQPIPHMLERNVPTMMSTAHAWQTILHGVQVLVSYMLMLVFMTYNTWLCAAVVLGSATGYFLFGWRESVVVDFTEHCH
Summary
Uniprot
Q2F612
H9JE94
A0A2A4K282
A0A3S2NMF5
S4PH06
I4DJU1
+ More
A0A194PJL6 A0A194RDL8 A0A212FH86 A0A1E1W982 A0A1E1VZV2 A0A2H1WFZ9 A0A1E1WQM7 A0A1E1WTZ0 A0A1E1WL17 A0A0C9QS65 A0A154P7E9 A0A0C9RE16 U5EZV7 A0A195BCV2 A0A158NIL0 E2BFD4 A0A1B6IF45 A0A232EY60 E2AVX5 A0A1B6HKH3 A0A0J7KXD9 A0A2J7PJ01 E0VNQ9 D6WCT1 A0A067R650 F4WPY2 N6T4U7 K7IRK0 A0A0P4W018 A0A1W4WS24 A0A1B6IPK9 A0A0L7R5F1 A0A1B6I9I9 R4G5Z0 A0A1J1HNR6 A0A2P8YLM4 Q16JJ6 A0A023EKM5 A0A088ACD2 A0A2A3EH69 W8BY43 V9IEK1 W8BMM3 V5GWU8 A0A0Q9W7H9 A0A1W4WS28 A0A023EKL4 A0A182F7Y9 A0A0K8V246 W5J979 A0A3R7N8U2 A0A2M3ZHU4 T1DSW0 A0A2M4BZT9 A0A0M9A8H4 A0A2M4BY30 A0A034VMC8 Q16JJ7 A0A0A1XHP6 A0A0R3NY01 A0A2M4AMD9 A0A2M4AL66 A0A3B0J967 W8C6V7 A0A0P4VLM0 R4FMH5 A0A0K8WJZ7 A0A0Q9XFG9 B4L5X4 A0A182G1Q8 A0A0M3QZJ9 A0A182H043 Q16JJ5 A0A0Q9WVR3 A0A1W4VSI3 A0A182QPS0 A0A069DP13 B4PYU1 C8VV93 A0A182VRE2
A0A194PJL6 A0A194RDL8 A0A212FH86 A0A1E1W982 A0A1E1VZV2 A0A2H1WFZ9 A0A1E1WQM7 A0A1E1WTZ0 A0A1E1WL17 A0A0C9QS65 A0A154P7E9 A0A0C9RE16 U5EZV7 A0A195BCV2 A0A158NIL0 E2BFD4 A0A1B6IF45 A0A232EY60 E2AVX5 A0A1B6HKH3 A0A0J7KXD9 A0A2J7PJ01 E0VNQ9 D6WCT1 A0A067R650 F4WPY2 N6T4U7 K7IRK0 A0A0P4W018 A0A1W4WS24 A0A1B6IPK9 A0A0L7R5F1 A0A1B6I9I9 R4G5Z0 A0A1J1HNR6 A0A2P8YLM4 Q16JJ6 A0A023EKM5 A0A088ACD2 A0A2A3EH69 W8BY43 V9IEK1 W8BMM3 V5GWU8 A0A0Q9W7H9 A0A1W4WS28 A0A023EKL4 A0A182F7Y9 A0A0K8V246 W5J979 A0A3R7N8U2 A0A2M3ZHU4 T1DSW0 A0A2M4BZT9 A0A0M9A8H4 A0A2M4BY30 A0A034VMC8 Q16JJ7 A0A0A1XHP6 A0A0R3NY01 A0A2M4AMD9 A0A2M4AL66 A0A3B0J967 W8C6V7 A0A0P4VLM0 R4FMH5 A0A0K8WJZ7 A0A0Q9XFG9 B4L5X4 A0A182G1Q8 A0A0M3QZJ9 A0A182H043 Q16JJ5 A0A0Q9WVR3 A0A1W4VSI3 A0A182QPS0 A0A069DP13 B4PYU1 C8VV93 A0A182VRE2
Pubmed
EMBL
DQ311260
ABD36205.1
BABH01002779
NWSH01000248
PCG78038.1
RSAL01000255
+ More
RVE43429.1 GAIX01005980 JAA86580.1 AK401559 BAM18181.1 KQ459602 KPI93228.1 KQ460367 KPJ15554.1 AGBW02008520 OWR53105.1 GDQN01007502 JAT83552.1 GDQN01010801 JAT80253.1 ODYU01008424 SOQ52015.1 GDQN01001908 JAT89146.1 GDQN01000748 JAT90306.1 GDQN01003503 JAT87551.1 GBYB01006594 JAG76361.1 KQ434831 KZC07807.1 GBYB01006595 JAG76362.1 GANO01000058 JAB59813.1 KQ976513 KYM82388.1 ADTU01016901 GL448009 EFN85594.1 GECU01035441 GECU01022147 GECU01018837 JAS72265.1 JAS85559.1 JAS88869.1 NNAY01001716 OXU23148.1 GL443213 EFN62397.1 GECU01032516 JAS75190.1 LBMM01002320 KMQ94986.1 NEVH01024952 PNF16283.1 DS235341 EEB15015.1 KQ971311 EEZ98850.1 KK852930 KDR13640.1 GL888257 EGI63740.1 APGK01052662 KB741213 ENN72648.1 GDRN01099761 JAI58715.1 GECU01036249 GECU01033103 GECU01023676 GECU01022288 GECU01021514 GECU01018851 JAS71457.1 JAS74603.1 JAS84030.1 JAS85418.1 JAS86192.1 JAS88855.1 KQ414654 KOC66049.1 GECU01030219 GECU01024105 GECU01009068 JAS77487.1 JAS83601.1 JAS98638.1 GAHY01000100 JAA77410.1 CVRI01000014 CRK89687.1 PYGN01000508 PSN45152.1 CH478006 EAT34445.1 GAPW01003882 JAC09716.1 KZ288256 PBC30632.1 GAMC01008324 JAB98231.1 JR039545 AEY58749.1 GAMC01008322 JAB98233.1 GALX01003718 JAB64748.1 CH940653 KRF80780.1 KRF80781.1 KRF80782.1 GAPW01003897 JAC09701.1 GDHF01019461 JAI32853.1 ADMH02002023 ETN59943.1 QCYY01001124 ROT80376.1 GGFM01007383 MBW28134.1 GAMD01001309 JAB00282.1 GGFJ01009422 MBW58563.1 KQ435712 KOX79390.1 GGFJ01008507 MBW57648.1 GAKP01014461 JAC44491.1 EAT34444.1 GBXI01006553 GBXI01003810 JAD07739.1 JAD10482.1 CH379063 KRT05832.1 KRT05833.1 KRT05834.1 GGFK01008619 MBW41940.1 GGFK01008215 MBW41536.1 OUUW01000003 SPP78804.1 GAMC01008326 JAB98229.1 GDKW01003026 JAI53569.1 ACPB03007038 GAHY01001714 JAA75796.1 GDHF01001129 JAI51185.1 CH933811 KRG07029.1 EDW06583.1 JXUM01038857 KQ561182 KXJ79454.1 CP012528 ALC49509.1 JXUM01100721 JXUM01100722 JXUM01100723 JXUM01100724 JXUM01100725 KQ564592 KXJ72044.1 EAT34446.1 CH964239 KRF99560.1 AXCN02000277 AXCN02000278 GBGD01003081 JAC85808.1 CM000162 EDX01027.1 KRK05897.1 KRK05898.1 KRK05899.1 KRK05901.1 BT099839 ACW83547.1
RVE43429.1 GAIX01005980 JAA86580.1 AK401559 BAM18181.1 KQ459602 KPI93228.1 KQ460367 KPJ15554.1 AGBW02008520 OWR53105.1 GDQN01007502 JAT83552.1 GDQN01010801 JAT80253.1 ODYU01008424 SOQ52015.1 GDQN01001908 JAT89146.1 GDQN01000748 JAT90306.1 GDQN01003503 JAT87551.1 GBYB01006594 JAG76361.1 KQ434831 KZC07807.1 GBYB01006595 JAG76362.1 GANO01000058 JAB59813.1 KQ976513 KYM82388.1 ADTU01016901 GL448009 EFN85594.1 GECU01035441 GECU01022147 GECU01018837 JAS72265.1 JAS85559.1 JAS88869.1 NNAY01001716 OXU23148.1 GL443213 EFN62397.1 GECU01032516 JAS75190.1 LBMM01002320 KMQ94986.1 NEVH01024952 PNF16283.1 DS235341 EEB15015.1 KQ971311 EEZ98850.1 KK852930 KDR13640.1 GL888257 EGI63740.1 APGK01052662 KB741213 ENN72648.1 GDRN01099761 JAI58715.1 GECU01036249 GECU01033103 GECU01023676 GECU01022288 GECU01021514 GECU01018851 JAS71457.1 JAS74603.1 JAS84030.1 JAS85418.1 JAS86192.1 JAS88855.1 KQ414654 KOC66049.1 GECU01030219 GECU01024105 GECU01009068 JAS77487.1 JAS83601.1 JAS98638.1 GAHY01000100 JAA77410.1 CVRI01000014 CRK89687.1 PYGN01000508 PSN45152.1 CH478006 EAT34445.1 GAPW01003882 JAC09716.1 KZ288256 PBC30632.1 GAMC01008324 JAB98231.1 JR039545 AEY58749.1 GAMC01008322 JAB98233.1 GALX01003718 JAB64748.1 CH940653 KRF80780.1 KRF80781.1 KRF80782.1 GAPW01003897 JAC09701.1 GDHF01019461 JAI32853.1 ADMH02002023 ETN59943.1 QCYY01001124 ROT80376.1 GGFM01007383 MBW28134.1 GAMD01001309 JAB00282.1 GGFJ01009422 MBW58563.1 KQ435712 KOX79390.1 GGFJ01008507 MBW57648.1 GAKP01014461 JAC44491.1 EAT34444.1 GBXI01006553 GBXI01003810 JAD07739.1 JAD10482.1 CH379063 KRT05832.1 KRT05833.1 KRT05834.1 GGFK01008619 MBW41940.1 GGFK01008215 MBW41536.1 OUUW01000003 SPP78804.1 GAMC01008326 JAB98229.1 GDKW01003026 JAI53569.1 ACPB03007038 GAHY01001714 JAA75796.1 GDHF01001129 JAI51185.1 CH933811 KRG07029.1 EDW06583.1 JXUM01038857 KQ561182 KXJ79454.1 CP012528 ALC49509.1 JXUM01100721 JXUM01100722 JXUM01100723 JXUM01100724 JXUM01100725 KQ564592 KXJ72044.1 EAT34446.1 CH964239 KRF99560.1 AXCN02000277 AXCN02000278 GBGD01003081 JAC85808.1 CM000162 EDX01027.1 KRK05897.1 KRK05898.1 KRK05899.1 KRK05901.1 BT099839 ACW83547.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000053240
UP000007151
+ More
UP000076502 UP000078540 UP000005205 UP000008237 UP000215335 UP000000311 UP000036403 UP000235965 UP000009046 UP000007266 UP000027135 UP000007755 UP000019118 UP000002358 UP000192223 UP000053825 UP000183832 UP000245037 UP000008820 UP000005203 UP000242457 UP000008792 UP000069272 UP000000673 UP000283509 UP000053105 UP000001819 UP000268350 UP000015103 UP000009192 UP000069940 UP000249989 UP000092553 UP000007798 UP000192221 UP000075886 UP000002282 UP000075920
UP000076502 UP000078540 UP000005205 UP000008237 UP000215335 UP000000311 UP000036403 UP000235965 UP000009046 UP000007266 UP000027135 UP000007755 UP000019118 UP000002358 UP000192223 UP000053825 UP000183832 UP000245037 UP000008820 UP000005203 UP000242457 UP000008792 UP000069272 UP000000673 UP000283509 UP000053105 UP000001819 UP000268350 UP000015103 UP000009192 UP000069940 UP000249989 UP000092553 UP000007798 UP000192221 UP000075886 UP000002282 UP000075920
Pfam
PF04145 Ctr
Interpro
IPR007274
Cop_transporter
ProteinModelPortal
Q2F612
H9JE94
A0A2A4K282
A0A3S2NMF5
S4PH06
I4DJU1
+ More
A0A194PJL6 A0A194RDL8 A0A212FH86 A0A1E1W982 A0A1E1VZV2 A0A2H1WFZ9 A0A1E1WQM7 A0A1E1WTZ0 A0A1E1WL17 A0A0C9QS65 A0A154P7E9 A0A0C9RE16 U5EZV7 A0A195BCV2 A0A158NIL0 E2BFD4 A0A1B6IF45 A0A232EY60 E2AVX5 A0A1B6HKH3 A0A0J7KXD9 A0A2J7PJ01 E0VNQ9 D6WCT1 A0A067R650 F4WPY2 N6T4U7 K7IRK0 A0A0P4W018 A0A1W4WS24 A0A1B6IPK9 A0A0L7R5F1 A0A1B6I9I9 R4G5Z0 A0A1J1HNR6 A0A2P8YLM4 Q16JJ6 A0A023EKM5 A0A088ACD2 A0A2A3EH69 W8BY43 V9IEK1 W8BMM3 V5GWU8 A0A0Q9W7H9 A0A1W4WS28 A0A023EKL4 A0A182F7Y9 A0A0K8V246 W5J979 A0A3R7N8U2 A0A2M3ZHU4 T1DSW0 A0A2M4BZT9 A0A0M9A8H4 A0A2M4BY30 A0A034VMC8 Q16JJ7 A0A0A1XHP6 A0A0R3NY01 A0A2M4AMD9 A0A2M4AL66 A0A3B0J967 W8C6V7 A0A0P4VLM0 R4FMH5 A0A0K8WJZ7 A0A0Q9XFG9 B4L5X4 A0A182G1Q8 A0A0M3QZJ9 A0A182H043 Q16JJ5 A0A0Q9WVR3 A0A1W4VSI3 A0A182QPS0 A0A069DP13 B4PYU1 C8VV93 A0A182VRE2
A0A194PJL6 A0A194RDL8 A0A212FH86 A0A1E1W982 A0A1E1VZV2 A0A2H1WFZ9 A0A1E1WQM7 A0A1E1WTZ0 A0A1E1WL17 A0A0C9QS65 A0A154P7E9 A0A0C9RE16 U5EZV7 A0A195BCV2 A0A158NIL0 E2BFD4 A0A1B6IF45 A0A232EY60 E2AVX5 A0A1B6HKH3 A0A0J7KXD9 A0A2J7PJ01 E0VNQ9 D6WCT1 A0A067R650 F4WPY2 N6T4U7 K7IRK0 A0A0P4W018 A0A1W4WS24 A0A1B6IPK9 A0A0L7R5F1 A0A1B6I9I9 R4G5Z0 A0A1J1HNR6 A0A2P8YLM4 Q16JJ6 A0A023EKM5 A0A088ACD2 A0A2A3EH69 W8BY43 V9IEK1 W8BMM3 V5GWU8 A0A0Q9W7H9 A0A1W4WS28 A0A023EKL4 A0A182F7Y9 A0A0K8V246 W5J979 A0A3R7N8U2 A0A2M3ZHU4 T1DSW0 A0A2M4BZT9 A0A0M9A8H4 A0A2M4BY30 A0A034VMC8 Q16JJ7 A0A0A1XHP6 A0A0R3NY01 A0A2M4AMD9 A0A2M4AL66 A0A3B0J967 W8C6V7 A0A0P4VLM0 R4FMH5 A0A0K8WJZ7 A0A0Q9XFG9 B4L5X4 A0A182G1Q8 A0A0M3QZJ9 A0A182H043 Q16JJ5 A0A0Q9WVR3 A0A1W4VSI3 A0A182QPS0 A0A069DP13 B4PYU1 C8VV93 A0A182VRE2
PDB
6M98
E-value=1.18039e-21,
Score=252
Ontologies
GO
PANTHER
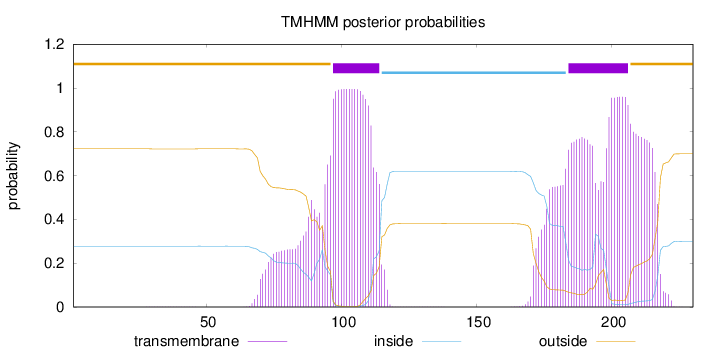
Topology
Length:
230
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
58.32334
Exp number, first 60 AAs:
0.00418
Total prob of N-in:
0.27766
outside
1 - 96
TMhelix
97 - 114
inside
115 - 183
TMhelix
184 - 206
outside
207 - 230
Population Genetic Test Statistics
Pi
242.456887
Theta
200.102899
Tajima's D
0.579053
CLR
0.274832
CSRT
0.542022898855057
Interpretation
Uncertain
