Pre Gene Modal
BGIBMGA007842
Annotation
PREDICTED:_protein_arginine_N-methyltransferase_5_isoform_X1_[Bombyx_mori]
Full name
Protein arginine N-methyltransferase
+ More
Protein arginine N-methyltransferase 5
Protein arginine N-methyltransferase 5
Location in the cell
Nuclear Reliability : 1.185
Sequence
CDS
ATGGCACAGCAAGAAATATCTTGTGGTTATGAATACATAATAACTGCTGATTTACAAACATGTCTCACAGAAGCACTTCAATGCAGTTATTCATTTATAGTTTCGCCCATTATTCACCCACGTTTTCGTAGGCAATCCACAAATGCCGGTAAAAATGGTGGTTTTACGCGGTCAGATATGGTTCTATCACCTCAAGATTGGACTAGCAGGATAGTAGCTAAGCTATCTCCGTACATTAACGTTGATTCACCTTCAGCGACTGTTAGGCAGCGTCACGAGGATTATTTGAATGAGGAACTATCATACTGTAGGGGACTTGGTGTGCCTGCAATCATGATATCAATACACGGGAGAGAGAGCAACAATTTAGCCAGAATTCTCCAAACTTATTATGAAACGAGCCACCATCCGTCTCTAATATGGGCATGTGTACCTATGCTATGTAGTAGAACATATAGAGAATGCACTGAAGATGATGAAGAAGAGAAAGCTTGGAATGAGCCCTGGTACTGGTGGTCCAAATTCCATGAGCGCCTTGATTGGGATAAACGTGTTGGTGTTGTTTTGGAGTTGTCTGCAGATCTTCCATCACAGGAAGTAGTAAAGAGATGGCTTGGTGAACCAGTAAAAGCGATTATCGTACCAACATCAATATTTCATAATAACAAAAAAGGATATCCTGTTTTGTCTCGGGCTCACCAACAACTTGTAGTTAGCATGGTAGAACATGAAGCACAGGTTATAGTTAGTGGTGCTCGAAGATCCAACATTGAATTCTTCGTACAGTATCTACACCGGGTGTGGAGAAGAAGGCCAAATCCAGCAAATGATCCTATGCTCAGCTATGCAAGAGGATGGGAAGACTACCTACAAACACCACTGCAGCCCCTAGCAGATAATTTAGATACACACACATATAATGTATTTGAAAAGGATCCAGTTAAATATAATCAATATCAAAAAGCTATTGCACAGGCGTTGATTGATGTACAGAAGGACCGGGCTGTGAAACAGATTGCTGAATATTTTCAGAGCAAAGCGAGCTGTAATGTAGCATCTATCAACCAATGTTCAAAAAATGAATTATGCGACAACAATACGAAAGAGTTGATTAGAGATGATCCTATAACTGTAATGGTACTTGGTGCGGGCCGGGGACCCCTAGTGAGAGCCACATTCAATGCTTCCGATATAACAAATACCAAAGTTAAGGTAATTGCCGTTGAGAAGAATCCATGTGCTGTAGTAGTGTTAGCTGCACAGGTACGAGAAGTCTGGCGTAATCGTGATGTGGTTGTTATACCTGGGGATATGAGGCAAATAAACTTGTCACCCAAAGCTGATATTATAGTTTCTGAATTGTTAGGTTCTTGGGGAGATAATGAGCTTTCGCCTGAATGCTTGGATGGAGCATCGAATCTTTTAAAACCCGGCGGAATATCGATTCCAAGCTCATATCGATCGTATGTTGCACCCATAACGTCTCCCAGGTTATGGGCTGCCGCGAAAATAGCAACTTCAGGAATTCCCCAACAAAAAGATAAGAACCTCGAAACATTATGGGTTGTTTACATGCAGAATAAACATGATATTGCAGAAACAAAGCTCGTCTTCACGTTTAACCATCCATCCAAAGCCGTAAAGGATGAAGAGGGATGTGATAAATCAGATTACAGAGGATTACCACTAACCAACAATAGGCGTCAGGCCACCCTTACTTGGGATGTTCAGCAAGACAACATTATGCATGGCTTTGGAGGGTATTTTGATTGTACCCTTTATGGTAATGAAATGATCAGCATTGTTCCTGGTACACATAGCCCAGGAATGATCTCCTGGTTTCCAGTCTTTATTCCAATTAAGACACCAATGCGAGTACAAAAGGGTGATAAAATAACAGCAACGTTCTGGAGGTGTGTTGACTCGCGTCGTGTATGGTACGAATGGGTAGTTGAAGTGGGTAACCGATCAACACCACTGCACAATGCTAACGGTCGTAGTTCGGAAATGTTGCTTTAA
Protein
MAQQEISCGYEYIITADLQTCLTEALQCSYSFIVSPIIHPRFRRQSTNAGKNGGFTRSDMVLSPQDWTSRIVAKLSPYINVDSPSATVRQRHEDYLNEELSYCRGLGVPAIMISIHGRESNNLARILQTYYETSHHPSLIWACVPMLCSRTYRECTEDDEEEKAWNEPWYWWSKFHERLDWDKRVGVVLELSADLPSQEVVKRWLGEPVKAIIVPTSIFHNNKKGYPVLSRAHQQLVVSMVEHEAQVIVSGARRSNIEFFVQYLHRVWRRRPNPANDPMLSYARGWEDYLQTPLQPLADNLDTHTYNVFEKDPVKYNQYQKAIAQALIDVQKDRAVKQIAEYFQSKASCNVASINQCSKNELCDNNTKELIRDDPITVMVLGAGRGPLVRATFNASDITNTKVKVIAVEKNPCAVVVLAAQVREVWRNRDVVVIPGDMRQINLSPKADIIVSELLGSWGDNELSPECLDGASNLLKPGGISIPSSYRSYVAPITSPRLWAAAKIATSGIPQQKDKNLETLWVVYMQNKHDIAETKLVFTFNHPSKAVKDEEGCDKSDYRGLPLTNNRRQATLTWDVQQDNIMHGFGGYFDCTLYGNEMISIVPGTHSPGMISWFPVFIPIKTPMRVQKGDKITATFWRCVDSRRVWYEWVVEVGNRSTPLHNANGRSSEMLL
Summary
Description
Arginine methyltransferase that can both catalyze the formation of omega-N monomethylarginine (MMA) and symmetrical dimethylarginine (sDMA).
Similarity
Belongs to the class I-like SAM-binding methyltransferase superfamily.
Belongs to the class I-like SAM-binding methyltransferase superfamily. Protein arginine N-methyltransferase family.
Belongs to the class I-like SAM-binding methyltransferase superfamily. Protein arginine N-methyltransferase family.
Uniprot
H9JE95
A0A2A4K2Y1
A0A194PII5
A0A194RDM7
A0A2H1WG18
S4PGH4
+ More
A0A212FHC1 A0A437AZ02 E0VNL3 R7V8B5 C3XU33 V5GU64 A0A131Y2Z5 A0A0M8ZTS8 A0A154P963 A0A098M1P2 H9G5L9 A0A1B6GLL8 A0A1B6IMB0 A0A1B6M2Q8 T1DNU4 H3CR85 A0A1A7WWK2 A0A1A7YT50 A0A3M6TRZ9 A0A1S3LZK4 H2RYB1 A0A3P8X1P0 A0A3B3SCG5 A0A060YB35 A0A1W5AL92 A0A1A8MG24 A0A1A8SKI0 A0A1A8PNI8 A0A3P8XG33 A0A1A8B2V4 V3ZAC9 A0A1A8U3W9 A0A1A8KVM2 A0A1A8FMM9 A0A1A8IL56 A0A0C9RQJ5 A0A1A8QNI1 A0A1A8GM81 A0A1A8CSY4 A0A3Q1C9B7 A0A3P8TT70 A0A3B5A1R2 A0A3Q3BNR2 A0A3B4GT08 W5LPJ2 I3KEK7 A0A0P4VU44 A0A3Q2Y7P4 A0A0B7AFA1 A0A1A8DFE0 A0A315V2K3 A0A3P9C2U5 T1HH53 A0A3Q1GI48 A0A126Q9B4 A0A3Q3N6U7 B1A4A0 A0A3Q2UW47 A0A3Q0RDX6 A0A3B3VU87 A0A087YG86 A0A0N8JYF4 H2MAB6 A0A3B3VS17 S7NMG4 A0A3B3X304 L5KL20 K9IUI7 A0A3B4CWQ1 A0A3P9H1A5 A0A060XTB2 G1PLM4 A0A147A2T7 A0A3P9LXG4 A0A3Q2PPR1 A0A2U9BKH8 A0A3B4UD77 A0A1S3R5B9 A0A341B735 M3ZE42 A0A3B4WA79 A0A2Y9E9V3 A0A2U3V0A8 A0A2Y9LKP8 A0A383ZZU9 H0WRB3 G3SV33 D2HTW0 K1PDE4 I3MFE3 A0A3Q7QDE4 A0A2Y9HGY0 A0A2U3W2C0
A0A212FHC1 A0A437AZ02 E0VNL3 R7V8B5 C3XU33 V5GU64 A0A131Y2Z5 A0A0M8ZTS8 A0A154P963 A0A098M1P2 H9G5L9 A0A1B6GLL8 A0A1B6IMB0 A0A1B6M2Q8 T1DNU4 H3CR85 A0A1A7WWK2 A0A1A7YT50 A0A3M6TRZ9 A0A1S3LZK4 H2RYB1 A0A3P8X1P0 A0A3B3SCG5 A0A060YB35 A0A1W5AL92 A0A1A8MG24 A0A1A8SKI0 A0A1A8PNI8 A0A3P8XG33 A0A1A8B2V4 V3ZAC9 A0A1A8U3W9 A0A1A8KVM2 A0A1A8FMM9 A0A1A8IL56 A0A0C9RQJ5 A0A1A8QNI1 A0A1A8GM81 A0A1A8CSY4 A0A3Q1C9B7 A0A3P8TT70 A0A3B5A1R2 A0A3Q3BNR2 A0A3B4GT08 W5LPJ2 I3KEK7 A0A0P4VU44 A0A3Q2Y7P4 A0A0B7AFA1 A0A1A8DFE0 A0A315V2K3 A0A3P9C2U5 T1HH53 A0A3Q1GI48 A0A126Q9B4 A0A3Q3N6U7 B1A4A0 A0A3Q2UW47 A0A3Q0RDX6 A0A3B3VU87 A0A087YG86 A0A0N8JYF4 H2MAB6 A0A3B3VS17 S7NMG4 A0A3B3X304 L5KL20 K9IUI7 A0A3B4CWQ1 A0A3P9H1A5 A0A060XTB2 G1PLM4 A0A147A2T7 A0A3P9LXG4 A0A3Q2PPR1 A0A2U9BKH8 A0A3B4UD77 A0A1S3R5B9 A0A341B735 M3ZE42 A0A3B4WA79 A0A2Y9E9V3 A0A2U3V0A8 A0A2Y9LKP8 A0A383ZZU9 H0WRB3 G3SV33 D2HTW0 K1PDE4 I3MFE3 A0A3Q7QDE4 A0A2Y9HGY0 A0A2U3W2C0
EC Number
2.1.1.-
Pubmed
EMBL
BABH01002781
NWSH01000248
PCG78040.1
KQ459602
KPI93226.1
KQ460367
+ More
KPJ15559.1 ODYU01008424 SOQ52013.1 GAIX01003667 JAA88893.1 AGBW02008520 OWR53107.1 RSAL01000255 RVE43432.1 DS235340 EEB14969.1 AMQN01004756 KB294299 ELU14784.1 GG666464 EEN68407.1 GANP01010553 JAB73915.1 GEFM01002941 JAP72855.1 KQ435891 KOX69369.1 KQ434846 KZC08382.1 GBSI01000265 JAC96230.1 AAWZ02033594 GECZ01006566 JAS63203.1 GECU01019671 JAS88035.1 GEBQ01009770 JAT30207.1 GAAZ01000207 JAA97736.1 HADW01008671 SBP10071.1 HADX01011515 SBP33747.1 RCHS01003049 RMX44172.1 FR907391 CDQ86609.1 HAEF01014716 SBR55875.1 HAEI01015939 SBS18408.1 HAEG01008856 SBR82970.1 HADY01022546 SBP61031.1 KB202823 ESO87913.1 HAEJ01001341 SBS41798.1 HAEE01015644 SBR35694.1 HAEB01012837 SBQ59364.1 HAED01010893 SBQ97158.1 GBYB01009494 GBYB01009496 JAG79261.1 JAG79263.1 HAEH01012467 SBR94848.1 HAEC01003427 SBQ71504.1 HADZ01017870 SBP81811.1 AERX01045779 GDKW01000424 JAI56171.1 HACG01032834 CEK79699.1 HAEA01003521 SBQ32001.1 NHOQ01002364 PWA17409.1 ACPB03009098 KT694369 AMK01821.1 EU419765 ABZ90976.1 AYCK01004988 JARO02005677 KPP66385.1 KE164377 EPQ17645.1 KB030709 ELK11258.1 GABZ01001168 JAA52357.1 FR905556 CDQ80150.1 AAPE02039928 GCES01013495 JAR72828.1 CP026249 AWP04578.1 AAQR03175737 AAQR03175738 AAQR03175739 AAQR03175740 GL193372 EFB18958.1 JH819184 EKC21902.1 AGTP01099390
KPJ15559.1 ODYU01008424 SOQ52013.1 GAIX01003667 JAA88893.1 AGBW02008520 OWR53107.1 RSAL01000255 RVE43432.1 DS235340 EEB14969.1 AMQN01004756 KB294299 ELU14784.1 GG666464 EEN68407.1 GANP01010553 JAB73915.1 GEFM01002941 JAP72855.1 KQ435891 KOX69369.1 KQ434846 KZC08382.1 GBSI01000265 JAC96230.1 AAWZ02033594 GECZ01006566 JAS63203.1 GECU01019671 JAS88035.1 GEBQ01009770 JAT30207.1 GAAZ01000207 JAA97736.1 HADW01008671 SBP10071.1 HADX01011515 SBP33747.1 RCHS01003049 RMX44172.1 FR907391 CDQ86609.1 HAEF01014716 SBR55875.1 HAEI01015939 SBS18408.1 HAEG01008856 SBR82970.1 HADY01022546 SBP61031.1 KB202823 ESO87913.1 HAEJ01001341 SBS41798.1 HAEE01015644 SBR35694.1 HAEB01012837 SBQ59364.1 HAED01010893 SBQ97158.1 GBYB01009494 GBYB01009496 JAG79261.1 JAG79263.1 HAEH01012467 SBR94848.1 HAEC01003427 SBQ71504.1 HADZ01017870 SBP81811.1 AERX01045779 GDKW01000424 JAI56171.1 HACG01032834 CEK79699.1 HAEA01003521 SBQ32001.1 NHOQ01002364 PWA17409.1 ACPB03009098 KT694369 AMK01821.1 EU419765 ABZ90976.1 AYCK01004988 JARO02005677 KPP66385.1 KE164377 EPQ17645.1 KB030709 ELK11258.1 GABZ01001168 JAA52357.1 FR905556 CDQ80150.1 AAPE02039928 GCES01013495 JAR72828.1 CP026249 AWP04578.1 AAQR03175737 AAQR03175738 AAQR03175739 AAQR03175740 GL193372 EFB18958.1 JH819184 EKC21902.1 AGTP01099390
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000283053
+ More
UP000009046 UP000014760 UP000001554 UP000053105 UP000076502 UP000001646 UP000007303 UP000275408 UP000087266 UP000005226 UP000265120 UP000261540 UP000193380 UP000192224 UP000265140 UP000030746 UP000257160 UP000265080 UP000261400 UP000264800 UP000261460 UP000018467 UP000005207 UP000264820 UP000265160 UP000015103 UP000257200 UP000261660 UP000264840 UP000261340 UP000261500 UP000028760 UP000034805 UP000001038 UP000261480 UP000010552 UP000261440 UP000265200 UP000001074 UP000265180 UP000265000 UP000246464 UP000261420 UP000252040 UP000002852 UP000261360 UP000248480 UP000245320 UP000248483 UP000261681 UP000005225 UP000007646 UP000005408 UP000005215 UP000286641 UP000248481 UP000245340
UP000009046 UP000014760 UP000001554 UP000053105 UP000076502 UP000001646 UP000007303 UP000275408 UP000087266 UP000005226 UP000265120 UP000261540 UP000193380 UP000192224 UP000265140 UP000030746 UP000257160 UP000265080 UP000261400 UP000264800 UP000261460 UP000018467 UP000005207 UP000264820 UP000265160 UP000015103 UP000257200 UP000261660 UP000264840 UP000261340 UP000261500 UP000028760 UP000034805 UP000001038 UP000261480 UP000010552 UP000261440 UP000265200 UP000001074 UP000265180 UP000265000 UP000246464 UP000261420 UP000252040 UP000002852 UP000261360 UP000248480 UP000245320 UP000248483 UP000261681 UP000005225 UP000007646 UP000005408 UP000005215 UP000286641 UP000248481 UP000245340
PRIDE
Interpro
SUPFAM
SSF53335
SSF53335
ProteinModelPortal
H9JE95
A0A2A4K2Y1
A0A194PII5
A0A194RDM7
A0A2H1WG18
S4PGH4
+ More
A0A212FHC1 A0A437AZ02 E0VNL3 R7V8B5 C3XU33 V5GU64 A0A131Y2Z5 A0A0M8ZTS8 A0A154P963 A0A098M1P2 H9G5L9 A0A1B6GLL8 A0A1B6IMB0 A0A1B6M2Q8 T1DNU4 H3CR85 A0A1A7WWK2 A0A1A7YT50 A0A3M6TRZ9 A0A1S3LZK4 H2RYB1 A0A3P8X1P0 A0A3B3SCG5 A0A060YB35 A0A1W5AL92 A0A1A8MG24 A0A1A8SKI0 A0A1A8PNI8 A0A3P8XG33 A0A1A8B2V4 V3ZAC9 A0A1A8U3W9 A0A1A8KVM2 A0A1A8FMM9 A0A1A8IL56 A0A0C9RQJ5 A0A1A8QNI1 A0A1A8GM81 A0A1A8CSY4 A0A3Q1C9B7 A0A3P8TT70 A0A3B5A1R2 A0A3Q3BNR2 A0A3B4GT08 W5LPJ2 I3KEK7 A0A0P4VU44 A0A3Q2Y7P4 A0A0B7AFA1 A0A1A8DFE0 A0A315V2K3 A0A3P9C2U5 T1HH53 A0A3Q1GI48 A0A126Q9B4 A0A3Q3N6U7 B1A4A0 A0A3Q2UW47 A0A3Q0RDX6 A0A3B3VU87 A0A087YG86 A0A0N8JYF4 H2MAB6 A0A3B3VS17 S7NMG4 A0A3B3X304 L5KL20 K9IUI7 A0A3B4CWQ1 A0A3P9H1A5 A0A060XTB2 G1PLM4 A0A147A2T7 A0A3P9LXG4 A0A3Q2PPR1 A0A2U9BKH8 A0A3B4UD77 A0A1S3R5B9 A0A341B735 M3ZE42 A0A3B4WA79 A0A2Y9E9V3 A0A2U3V0A8 A0A2Y9LKP8 A0A383ZZU9 H0WRB3 G3SV33 D2HTW0 K1PDE4 I3MFE3 A0A3Q7QDE4 A0A2Y9HGY0 A0A2U3W2C0
A0A212FHC1 A0A437AZ02 E0VNL3 R7V8B5 C3XU33 V5GU64 A0A131Y2Z5 A0A0M8ZTS8 A0A154P963 A0A098M1P2 H9G5L9 A0A1B6GLL8 A0A1B6IMB0 A0A1B6M2Q8 T1DNU4 H3CR85 A0A1A7WWK2 A0A1A7YT50 A0A3M6TRZ9 A0A1S3LZK4 H2RYB1 A0A3P8X1P0 A0A3B3SCG5 A0A060YB35 A0A1W5AL92 A0A1A8MG24 A0A1A8SKI0 A0A1A8PNI8 A0A3P8XG33 A0A1A8B2V4 V3ZAC9 A0A1A8U3W9 A0A1A8KVM2 A0A1A8FMM9 A0A1A8IL56 A0A0C9RQJ5 A0A1A8QNI1 A0A1A8GM81 A0A1A8CSY4 A0A3Q1C9B7 A0A3P8TT70 A0A3B5A1R2 A0A3Q3BNR2 A0A3B4GT08 W5LPJ2 I3KEK7 A0A0P4VU44 A0A3Q2Y7P4 A0A0B7AFA1 A0A1A8DFE0 A0A315V2K3 A0A3P9C2U5 T1HH53 A0A3Q1GI48 A0A126Q9B4 A0A3Q3N6U7 B1A4A0 A0A3Q2UW47 A0A3Q0RDX6 A0A3B3VU87 A0A087YG86 A0A0N8JYF4 H2MAB6 A0A3B3VS17 S7NMG4 A0A3B3X304 L5KL20 K9IUI7 A0A3B4CWQ1 A0A3P9H1A5 A0A060XTB2 G1PLM4 A0A147A2T7 A0A3P9LXG4 A0A3Q2PPR1 A0A2U9BKH8 A0A3B4UD77 A0A1S3R5B9 A0A341B735 M3ZE42 A0A3B4WA79 A0A2Y9E9V3 A0A2U3V0A8 A0A2Y9LKP8 A0A383ZZU9 H0WRB3 G3SV33 D2HTW0 K1PDE4 I3MFE3 A0A3Q7QDE4 A0A2Y9HGY0 A0A2U3W2C0
PDB
6CKC
E-value=3.17073e-141,
Score=1288
Ontologies
GO
GO:0008168
GO:0035246
GO:0008469
GO:0005829
GO:0005634
GO:0034969
GO:0016301
GO:0044020
GO:0005794
GO:0042118
GO:0035243
GO:0035097
GO:0090161
GO:0046982
GO:0008327
GO:0042802
GO:0043021
GO:0002039
GO:1904992
GO:0000387
GO:0006353
GO:0034709
GO:0048742
GO:0006479
GO:0008270
GO:0015074
GO:0016817
GO:0043565
GO:0016876
GO:0043039
PANTHER
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleus
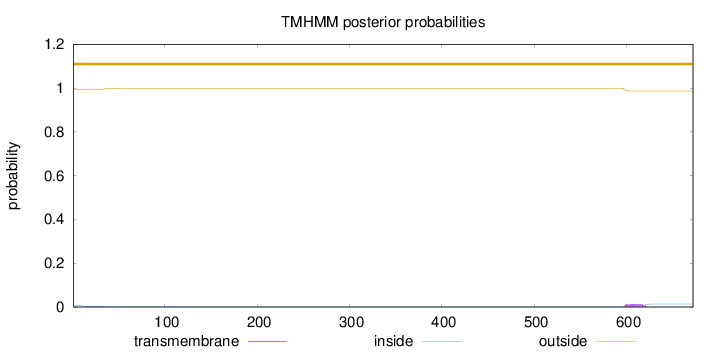
Length:
672
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.38487
Exp number, first 60 AAs:
0.09154
Total prob of N-in:
0.00546
outside
1 - 672
Population Genetic Test Statistics
Pi
245.097288
Theta
182.776842
Tajima's D
1.087679
CLR
0.003529
CSRT
0.692065396730164
Interpretation
Uncertain
