Pre Gene Modal
BGIBMGA007845
Annotation
putative_mitochondrial_ribosomal_protein?_L51_[Operophtera_brumata]
Full name
39S ribosomal protein L51, mitochondrial
Location in the cell
Mitochondrial Reliability : 3.692
Sequence
CDS
ATGAGCTGGTTAACTTCGAGGATTCTAAGCGTTGTTTGGAAACCCACAGTAAACGTTGTAAGGACAAGATATCACGCCGATAAAAAACGATTAGTCAGGAGATATGGCTACCAAGAGAAAATCTGGCGTGGTGGACTCTTGCCTAGATCTGAAGGAAAGGCACTACCAATACCTGAATACAGACCTGATAATCCTTGGTCAGTAAGAAAAGCTCTTTTTGGACAAAATGACTATATAGACATTTTAGGACCTGGTAATCTACATCCAGTTAAAACATTATATAATATACCGTCATGGATTCGGGGTGTGAGTGGACATGAATATCATATCCTACTAAGAAAAAAGAAAATGTGGGCAAAAACTGGTACACCTAGATTGAGACCTACAAAATGGAAGGAAATACAAAAGCGCTTGTCGTTCCTGTATAGAAAACTAAATAGAAAAACGAAGACAGGACCTTCACCAATTTAA
Protein
MSWLTSRILSVVWKPTVNVVRTRYHADKKRLVRRYGYQEKIWRGGLLPRSEGKALPIPEYRPDNPWSVRKALFGQNDYIDILGPGNLHPVKTLYNIPSWIRGVSGHEYHILLRKKKMWAKTGTPRLRPTKWKEIQKRLSFLYRKLNRKTKTGPSPI
Summary
Subunit
Component of the mitochondrial ribosome large subunit (39S) which comprises a 16S rRNA and about 50 distinct proteins (By similarity).
Similarity
Belongs to the mitochondrion-specific ribosomal protein mL51 family.
Keywords
Complete proteome
Mitochondrion
Reference proteome
Ribonucleoprotein
Ribosomal protein
Transit peptide
Feature
chain 39S ribosomal protein L51, mitochondrial
Uniprot
H9JE98
A0A0L7L4R7
A0A194PJL0
A0A194RD78
A0A2H1WG06
A0A437B0E1
+ More
A0A2A4K1C5 A0A212FHA8 A0A023EG54 Q16TN5 B0WUM9 A0A310ST77 A0A1Q3FFB5 A0A182VXX5 A0A182IPL3 A0A182N4G7 A0A182WV87 A0A182PZG0 A0A182I8W1 A0A182UBB8 Q7PS66 A0A182VAV8 A0A182M8J2 A0A182R5U3 A0A182XXG1 A0A182P7A7 A0A182JZR9 A0A084VGL9 A0A2M4C1P3 A0A2M3ZC57 A0A2M4C1Q8 A0A2M4AP69 A0A2M4C1P1 W5JA05 A0A0N0BDH8 A0A182F7N7 A0A2M4APF0 A0A195FGV0 A0A151WER4 A0A2M3ZJ71 E9IHK4 A0A151JNB9 A0A158NJ39 A0A0J7L6M5 U5EGB8 B4LRC0 A0A067R5Z5 A0A0L7RHT8 A0A1B0CZT9 T1DNV8 B4Q6V8 F4X389 A0A088AQ53 B4HYN9 E2BNG5 Q9VLJ9 A0A151IL01 A0A2J7Q4P6 B4KI54 B4NXF6 A0A026WEK2 B3MLX6 D3TP57 A0A1Q3FFB7 B4GKQ2 Q29PG4 B5DS73 B3N7C3 A0A1J1J2A5 E2AM61 A0A1I8PIS0 A0A1I8M9R0 A0A2P8Z0W9 B4N1B6 A0A1L8E158 A0A1A9ZNU1 A0A1A9VBD5 A0A1A9Y7F1 A0A1I8PJ01 A0A3B0JZR6 A0A1L8E1J4 A0A2A3EAL3 A0A1Y1MB16 T1HV86 A0A1B0C3G7 A0A336M0V8 A0A1B0C9J7 A0A1W4W8Q0 A0A0A1WE18 A0A0L0BZ05 A0A034WQK9 B4JBC3 A0A0K8UZF8 A0A0K8UJD4 A0A0V0G9F6 A0A0M4E8F5 A0A146L2M4 A0A069DW25 A0A0A9WW05 V5GCR9
A0A2A4K1C5 A0A212FHA8 A0A023EG54 Q16TN5 B0WUM9 A0A310ST77 A0A1Q3FFB5 A0A182VXX5 A0A182IPL3 A0A182N4G7 A0A182WV87 A0A182PZG0 A0A182I8W1 A0A182UBB8 Q7PS66 A0A182VAV8 A0A182M8J2 A0A182R5U3 A0A182XXG1 A0A182P7A7 A0A182JZR9 A0A084VGL9 A0A2M4C1P3 A0A2M3ZC57 A0A2M4C1Q8 A0A2M4AP69 A0A2M4C1P1 W5JA05 A0A0N0BDH8 A0A182F7N7 A0A2M4APF0 A0A195FGV0 A0A151WER4 A0A2M3ZJ71 E9IHK4 A0A151JNB9 A0A158NJ39 A0A0J7L6M5 U5EGB8 B4LRC0 A0A067R5Z5 A0A0L7RHT8 A0A1B0CZT9 T1DNV8 B4Q6V8 F4X389 A0A088AQ53 B4HYN9 E2BNG5 Q9VLJ9 A0A151IL01 A0A2J7Q4P6 B4KI54 B4NXF6 A0A026WEK2 B3MLX6 D3TP57 A0A1Q3FFB7 B4GKQ2 Q29PG4 B5DS73 B3N7C3 A0A1J1J2A5 E2AM61 A0A1I8PIS0 A0A1I8M9R0 A0A2P8Z0W9 B4N1B6 A0A1L8E158 A0A1A9ZNU1 A0A1A9VBD5 A0A1A9Y7F1 A0A1I8PJ01 A0A3B0JZR6 A0A1L8E1J4 A0A2A3EAL3 A0A1Y1MB16 T1HV86 A0A1B0C3G7 A0A336M0V8 A0A1B0C9J7 A0A1W4W8Q0 A0A0A1WE18 A0A0L0BZ05 A0A034WQK9 B4JBC3 A0A0K8UZF8 A0A0K8UJD4 A0A0V0G9F6 A0A0M4E8F5 A0A146L2M4 A0A069DW25 A0A0A9WW05 V5GCR9
Pubmed
19121390
26227816
26354079
22118469
24945155
26483478
+ More
17510324 12364791 14747013 17210077 25244985 24438588 20920257 23761445 21282665 21347285 17994087 24845553 22936249 21719571 20798317 10731132 12537572 12537569 17550304 24508170 30249741 20353571 15632085 25315136 29403074 28004739 25830018 26108605 25348373 26823975 26334808 25401762
17510324 12364791 14747013 17210077 25244985 24438588 20920257 23761445 21282665 21347285 17994087 24845553 22936249 21719571 20798317 10731132 12537572 12537569 17550304 24508170 30249741 20353571 15632085 25315136 29403074 28004739 25830018 26108605 25348373 26823975 26334808 25401762
EMBL
BABH01002787
BABH01002788
JTDY01003005
KOB70326.1
KQ459602
KPI93223.1
+ More
KQ460367 KPJ15562.1 ODYU01008424 SOQ52009.1 RSAL01000230 RVE43854.1 NWSH01000248 PCG78047.1 AGBW02008520 OWR53110.1 JXUM01042299 GAPW01005220 KQ561318 JAC08378.1 KXJ79011.1 CH477643 EAT37895.1 DS232107 EDS35014.1 KQ760478 OAD60217.1 GFDL01008800 JAV26245.1 AXCN02000040 APCN01003126 AAAB01008844 EAA06096.4 AXCM01007023 ATLV01012999 KE524833 KFB37113.1 GGFJ01010099 MBW59240.1 GGFM01005304 MBW26055.1 GGFJ01010098 MBW59239.1 GGFK01009260 MBW42581.1 GGFJ01010091 MBW59232.1 ADMH02001961 ETN60243.1 KQ435876 KOX70114.1 GGFK01009328 MBW42649.1 KQ981606 KYN39601.1 KQ983238 KYQ46271.1 GGFM01007835 MBW28586.1 GL763289 EFZ19957.1 KQ978904 KYN27553.1 ADTU01000267 LBMM01000449 KMQ98316.1 GANO01003509 JAB56362.1 CH940649 EDW64590.1 KK897593 KK852672 KDQ65279.1 KDR18779.1 KQ414590 KOC70379.1 AJVK01009723 GAMD01002525 JAA99065.1 CM000361 CM002910 EDX04267.1 KMY89120.1 GL888609 EGI59070.1 CH480818 EDW52169.1 GL449414 EFN82744.1 AE014134 AY051652 KQ977151 KYN05269.1 NEVH01018378 PNF23553.1 CH933807 EDW12347.1 CM000157 EDW88547.1 KK107250 QOIP01000002 EZA54378.1 RLU25446.1 CH902620 EDV31804.1 CCAG010000321 EZ423209 ADD19485.1 GFDL01008796 JAV26249.1 CH479184 EDW37218.1 CH379058 CH475461 EDY71204.1 CH954177 EDV58274.2 CVRI01000067 CRL06519.1 GL440715 EFN65467.1 PYGN01000249 PSN50156.1 CH963920 EDW78126.1 GFDF01001653 JAV12431.1 OUUW01000010 SPP85912.1 GFDF01001652 JAV12432.1 KZ288320 PBC28249.1 GEZM01036337 GEZM01036336 GEZM01036335 GEZM01036334 GEZM01036333 GEZM01036332 GEZM01036331 GEZM01036330 GEZM01036329 GEZM01036328 GEZM01036327 GEZM01036326 JAV82801.1 ACPB03025846 JXJN01024951 UFQT01000257 SSX22473.1 AJWK01002423 GBXI01017624 JAC96667.1 JRES01001227 KNC24469.1 GAKP01002345 JAC56607.1 CH916368 EDW02928.1 GDHF01020257 JAI32057.1 GDHF01025638 JAI26676.1 GECL01001486 JAP04638.1 CP012523 ALC38276.1 GDHC01015835 JAQ02794.1 GBGD01003370 JAC85519.1 GBHO01030982 GBRD01005416 JAG12622.1 JAG60405.1 GALX01006642 JAB61824.1
KQ460367 KPJ15562.1 ODYU01008424 SOQ52009.1 RSAL01000230 RVE43854.1 NWSH01000248 PCG78047.1 AGBW02008520 OWR53110.1 JXUM01042299 GAPW01005220 KQ561318 JAC08378.1 KXJ79011.1 CH477643 EAT37895.1 DS232107 EDS35014.1 KQ760478 OAD60217.1 GFDL01008800 JAV26245.1 AXCN02000040 APCN01003126 AAAB01008844 EAA06096.4 AXCM01007023 ATLV01012999 KE524833 KFB37113.1 GGFJ01010099 MBW59240.1 GGFM01005304 MBW26055.1 GGFJ01010098 MBW59239.1 GGFK01009260 MBW42581.1 GGFJ01010091 MBW59232.1 ADMH02001961 ETN60243.1 KQ435876 KOX70114.1 GGFK01009328 MBW42649.1 KQ981606 KYN39601.1 KQ983238 KYQ46271.1 GGFM01007835 MBW28586.1 GL763289 EFZ19957.1 KQ978904 KYN27553.1 ADTU01000267 LBMM01000449 KMQ98316.1 GANO01003509 JAB56362.1 CH940649 EDW64590.1 KK897593 KK852672 KDQ65279.1 KDR18779.1 KQ414590 KOC70379.1 AJVK01009723 GAMD01002525 JAA99065.1 CM000361 CM002910 EDX04267.1 KMY89120.1 GL888609 EGI59070.1 CH480818 EDW52169.1 GL449414 EFN82744.1 AE014134 AY051652 KQ977151 KYN05269.1 NEVH01018378 PNF23553.1 CH933807 EDW12347.1 CM000157 EDW88547.1 KK107250 QOIP01000002 EZA54378.1 RLU25446.1 CH902620 EDV31804.1 CCAG010000321 EZ423209 ADD19485.1 GFDL01008796 JAV26249.1 CH479184 EDW37218.1 CH379058 CH475461 EDY71204.1 CH954177 EDV58274.2 CVRI01000067 CRL06519.1 GL440715 EFN65467.1 PYGN01000249 PSN50156.1 CH963920 EDW78126.1 GFDF01001653 JAV12431.1 OUUW01000010 SPP85912.1 GFDF01001652 JAV12432.1 KZ288320 PBC28249.1 GEZM01036337 GEZM01036336 GEZM01036335 GEZM01036334 GEZM01036333 GEZM01036332 GEZM01036331 GEZM01036330 GEZM01036329 GEZM01036328 GEZM01036327 GEZM01036326 JAV82801.1 ACPB03025846 JXJN01024951 UFQT01000257 SSX22473.1 AJWK01002423 GBXI01017624 JAC96667.1 JRES01001227 KNC24469.1 GAKP01002345 JAC56607.1 CH916368 EDW02928.1 GDHF01020257 JAI32057.1 GDHF01025638 JAI26676.1 GECL01001486 JAP04638.1 CP012523 ALC38276.1 GDHC01015835 JAQ02794.1 GBGD01003370 JAC85519.1 GBHO01030982 GBRD01005416 JAG12622.1 JAG60405.1 GALX01006642 JAB61824.1
Proteomes
UP000005204
UP000037510
UP000053268
UP000053240
UP000283053
UP000218220
+ More
UP000007151 UP000069940 UP000249989 UP000008820 UP000002320 UP000075920 UP000075880 UP000075884 UP000076407 UP000075886 UP000075840 UP000075902 UP000007062 UP000075903 UP000075883 UP000075900 UP000076408 UP000075885 UP000075881 UP000030765 UP000000673 UP000053105 UP000069272 UP000078541 UP000075809 UP000078492 UP000005205 UP000036403 UP000008792 UP000027135 UP000053825 UP000092462 UP000000304 UP000007755 UP000005203 UP000001292 UP000008237 UP000000803 UP000078542 UP000235965 UP000009192 UP000002282 UP000053097 UP000279307 UP000007801 UP000092444 UP000008744 UP000001819 UP000008711 UP000183832 UP000000311 UP000095300 UP000095301 UP000245037 UP000007798 UP000092445 UP000078200 UP000092443 UP000268350 UP000242457 UP000015103 UP000092460 UP000092461 UP000192221 UP000037069 UP000001070 UP000092553
UP000007151 UP000069940 UP000249989 UP000008820 UP000002320 UP000075920 UP000075880 UP000075884 UP000076407 UP000075886 UP000075840 UP000075902 UP000007062 UP000075903 UP000075883 UP000075900 UP000076408 UP000075885 UP000075881 UP000030765 UP000000673 UP000053105 UP000069272 UP000078541 UP000075809 UP000078492 UP000005205 UP000036403 UP000008792 UP000027135 UP000053825 UP000092462 UP000000304 UP000007755 UP000005203 UP000001292 UP000008237 UP000000803 UP000078542 UP000235965 UP000009192 UP000002282 UP000053097 UP000279307 UP000007801 UP000092444 UP000008744 UP000001819 UP000008711 UP000183832 UP000000311 UP000095300 UP000095301 UP000245037 UP000007798 UP000092445 UP000078200 UP000092443 UP000268350 UP000242457 UP000015103 UP000092460 UP000092461 UP000192221 UP000037069 UP000001070 UP000092553
Pfam
PF10244 MRP-L51
Interpro
IPR019373
Ribosomal_L51_mit
ProteinModelPortal
H9JE98
A0A0L7L4R7
A0A194PJL0
A0A194RD78
A0A2H1WG06
A0A437B0E1
+ More
A0A2A4K1C5 A0A212FHA8 A0A023EG54 Q16TN5 B0WUM9 A0A310ST77 A0A1Q3FFB5 A0A182VXX5 A0A182IPL3 A0A182N4G7 A0A182WV87 A0A182PZG0 A0A182I8W1 A0A182UBB8 Q7PS66 A0A182VAV8 A0A182M8J2 A0A182R5U3 A0A182XXG1 A0A182P7A7 A0A182JZR9 A0A084VGL9 A0A2M4C1P3 A0A2M3ZC57 A0A2M4C1Q8 A0A2M4AP69 A0A2M4C1P1 W5JA05 A0A0N0BDH8 A0A182F7N7 A0A2M4APF0 A0A195FGV0 A0A151WER4 A0A2M3ZJ71 E9IHK4 A0A151JNB9 A0A158NJ39 A0A0J7L6M5 U5EGB8 B4LRC0 A0A067R5Z5 A0A0L7RHT8 A0A1B0CZT9 T1DNV8 B4Q6V8 F4X389 A0A088AQ53 B4HYN9 E2BNG5 Q9VLJ9 A0A151IL01 A0A2J7Q4P6 B4KI54 B4NXF6 A0A026WEK2 B3MLX6 D3TP57 A0A1Q3FFB7 B4GKQ2 Q29PG4 B5DS73 B3N7C3 A0A1J1J2A5 E2AM61 A0A1I8PIS0 A0A1I8M9R0 A0A2P8Z0W9 B4N1B6 A0A1L8E158 A0A1A9ZNU1 A0A1A9VBD5 A0A1A9Y7F1 A0A1I8PJ01 A0A3B0JZR6 A0A1L8E1J4 A0A2A3EAL3 A0A1Y1MB16 T1HV86 A0A1B0C3G7 A0A336M0V8 A0A1B0C9J7 A0A1W4W8Q0 A0A0A1WE18 A0A0L0BZ05 A0A034WQK9 B4JBC3 A0A0K8UZF8 A0A0K8UJD4 A0A0V0G9F6 A0A0M4E8F5 A0A146L2M4 A0A069DW25 A0A0A9WW05 V5GCR9
A0A2A4K1C5 A0A212FHA8 A0A023EG54 Q16TN5 B0WUM9 A0A310ST77 A0A1Q3FFB5 A0A182VXX5 A0A182IPL3 A0A182N4G7 A0A182WV87 A0A182PZG0 A0A182I8W1 A0A182UBB8 Q7PS66 A0A182VAV8 A0A182M8J2 A0A182R5U3 A0A182XXG1 A0A182P7A7 A0A182JZR9 A0A084VGL9 A0A2M4C1P3 A0A2M3ZC57 A0A2M4C1Q8 A0A2M4AP69 A0A2M4C1P1 W5JA05 A0A0N0BDH8 A0A182F7N7 A0A2M4APF0 A0A195FGV0 A0A151WER4 A0A2M3ZJ71 E9IHK4 A0A151JNB9 A0A158NJ39 A0A0J7L6M5 U5EGB8 B4LRC0 A0A067R5Z5 A0A0L7RHT8 A0A1B0CZT9 T1DNV8 B4Q6V8 F4X389 A0A088AQ53 B4HYN9 E2BNG5 Q9VLJ9 A0A151IL01 A0A2J7Q4P6 B4KI54 B4NXF6 A0A026WEK2 B3MLX6 D3TP57 A0A1Q3FFB7 B4GKQ2 Q29PG4 B5DS73 B3N7C3 A0A1J1J2A5 E2AM61 A0A1I8PIS0 A0A1I8M9R0 A0A2P8Z0W9 B4N1B6 A0A1L8E158 A0A1A9ZNU1 A0A1A9VBD5 A0A1A9Y7F1 A0A1I8PJ01 A0A3B0JZR6 A0A1L8E1J4 A0A2A3EAL3 A0A1Y1MB16 T1HV86 A0A1B0C3G7 A0A336M0V8 A0A1B0C9J7 A0A1W4W8Q0 A0A0A1WE18 A0A0L0BZ05 A0A034WQK9 B4JBC3 A0A0K8UZF8 A0A0K8UJD4 A0A0V0G9F6 A0A0M4E8F5 A0A146L2M4 A0A069DW25 A0A0A9WW05 V5GCR9
PDB
6NU3
E-value=3.70821e-07,
Score=124
Ontologies
GO
PANTHER
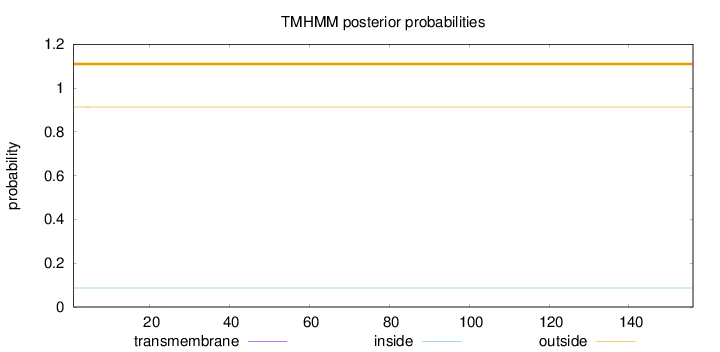
Topology
Subcellular location
Mitochondrion
Length:
156
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01119
Exp number, first 60 AAs:
0.00921
Total prob of N-in:
0.08651
outside
1 - 156
Population Genetic Test Statistics
Pi
326.944099
Theta
226.247276
Tajima's D
1.522237
CLR
0
CSRT
0.795010249487526
Interpretation
Uncertain
