Gene
KWMTBOMO08858 Validated by peptides from experiments
Annotation
cytochrome_P450_CYP332A1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.256 PlasmaMembrane Reliability : 1.604
Sequence
CDS
ATGGTCGATGTAAACTCTTATACAATTTACCTGTTGTTCAATCGGAATTATCAATACTGGGAGAAACGGGGCGTGCCTTACGAAAAGCCGTTCTTCTTGTTCGGAAGTTTGTCTTTTATCCTGAGGAAAAGCTTTTGGGACTATTTCTACGAACTAAGCAAGAGGCATACGGGTGACTATGTTGGAATATTTTTGGGATTCAAACCGACCCTTATGGTACAGACTCCAGAAATAGCGCGTCGTATCCTGGTCAAGGACAATGCCCATTTCAACAACCGATACTGTTATTCTTCGTACGGCGTGGATCCACTGGGCTCCTTAAATTTGTTTACAGTAAAGAATCCGAAATGGAGCAATATCAGACATGAGCTATCTCCGATGTTCACTTCGTTACGACTGAAGACGATATGTGAACTGATGAACGTTAACGCTAAAGAGCTTGTATTGAAAATTCAGCGAGATTACATTGATAATAATGAAGACGTCAACTTGAAGGAATTGTTCTCAATGTACACATCGGATACTGTAGGTTATACAGTATTTGGTCTCCGCGTGAGTGCACTCAATGATCCGTCCTCGCCTCTCTGGTTCATCACTAACCACATGGTCAAGTGGGACTTTTGGCGCGGCTTCGAATTCACAGCCATCTTCTTTGTTCCTGCACTAGCTCGTTTCTTAAGGTTAAAATTCTTTTCACAACCGGCCACTGAATATATTATGAGACTATTCCGGACCGTTGTCGATGAACGCAAAAAGACGAACCAGAACACCGATAAGGACCTTGTTAACCATTTGCTGAAGCTGAAAGAAAATTTGAAGCTTGGAGCTGATATAAAGCTCGCCGACGAGATAATGATGGCGCAAGCAGCTGTATTCATTTTAGGTTCCATAGAAACATCATCATCAACTCTGTCTTACTGTCTGCACGAGTTAGCGTACCATCCCGAAGAACAGCAAAAACTGTTTGAGGAAGTGGATGACGCAATAAAGGAGACCGGAAAAGAAATTCTAGACTACGAAAATTTACAAGAACTCAAATATCTAAGCGCTTGCATTCTGGAAACTTTGCGCAAATATCCTCCGGTGCCGCACATAGATAGAGTTTGTAACAAAACATACAAACTAAACGATGAATTAACAATAGAAGAGGATATACCGGTGTTCGTGAATGTACTGGCGATCCATCGCAACGAGAAATACTACCCTGAGCCGGACCAATGGCGTCCTGAGAGAATGATCGGGGTCACTGACAATGACAATTTGCAGTATACGTTCCTGCCCTTCGGAGATGGGCCACGTTTTTGCATAGGAAAACGTTACGGTTTGTTGCAAATGAGGGCCGCCATTGCCCAGATGATTCATAAATACAAATTCGAAGCTGCGGAACCACACAGCACTCCGAGCGACCCGTACAGCGTCATCCTGTCACCTAAATCAGGTGGCCGTATTAAATTCGTACCGCGATAA
Protein
MVDVNSYTIYLLFNRNYQYWEKRGVPYEKPFFLFGSLSFILRKSFWDYFYELSKRHTGDYVGIFLGFKPTLMVQTPEIARRILVKDNAHFNNRYCYSSYGVDPLGSLNLFTVKNPKWSNIRHELSPMFTSLRLKTICELMNVNAKELVLKIQRDYIDNNEDVNLKELFSMYTSDTVGYTVFGLRVSALNDPSSPLWFITNHMVKWDFWRGFEFTAIFFVPALARFLRLKFFSQPATEYIMRLFRTVVDERKKTNQNTDKDLVNHLLKLKENLKLGADIKLADEIMMAQAAVFILGSIETSSSTLSYCLHELAYHPEEQQKLFEEVDDAIKETGKEILDYENLQELKYLSACILETLRKYPPVPHIDRVCNKTYKLNDELTIEEDIPVFVNVLAIHRNEKYYPEPDQWRPERMIGVTDNDNLQYTFLPFGDGPRFCIGKRYGLLQMRAAIAQMIHKYKFEAAEPHSTPSDPYSVILSPKSGGRIKFVPR
Summary
Cofactor
heme
Similarity
Belongs to the cytochrome P450 family.
Uniprot
B1AAB5
A0A2Z6JJT5
D5L0N3
A0A2Z5CUB7
A0A2Z6JI91
A0A248QFE5
+ More
D5L0N2 A0A2Z6JJS6 A0A2A4K2A3 A0A2S1PSQ3 A0A2Z5CT64 A0A068F0U3 A0A2S1PSN1 A0A2Z5CT56 A0A2Z6JKL5 A0A1V0D9H5 A0A2Z5CT65 A0A2Z5CT45 A0A2Z6JKK7 A0A2Z5CTX4 A0A2Z6JQN4 A0A2Z6JSP9 A0A2Z5CVH7 A0A0K8TVQ8 A0A2Z6JKK5 A0A2Z6JIS5 A0A2Z6JIS9 A0A2Z6JJ49 A0A2Z6JVD1 A0A2Z5CVH9 A0A2Z5CT48 A0A2Z6JNK9 A0A2Z6JLZ3 A0A172MFA5 A0A2Z6JNK2 A0A2Z6JSR4 A0A2Z6JIS3 A0A2Z6JNJ2 A0A2Z6JIS1 A0A2Z5CV12 A0A2Z6JLY7 A0A1E1WI40 A0A2Z6JQP2 A0A2Z6JVD9 A0A2Z5CV54 A0A2Z5CTZ2 A0A2Z5CT49 A0A2Z5CTW1 A0A194RE58 A0A2Z6JJS2 A0A194PQ11 A0A2A4K2N2 D2JLJ8 A0A212FGZ1 A0A2Z5CU05 A0A2Z6JI71 A0A1V0D9G7 A0A0C5C563 A0A1V0D9G8 A0A2Z5CV07 A0A1V0D9G1 A0A1V0D9H1 A0A212ESQ5 A0A1V0D9G0 A0A068EU25 A0A3S2NRQ2 A0A194RM59 A0A194QLW1 A0A2W1BF80 A0A2H1V767 A0A2H1V738 A0A3G5EAS2 A0A2W1BMN3 A0A2A4JVD4 A0A0M9A0V9 A0A212FBK5 A0A286QUF5 A0A1W6L1S3 A0A248QJ37 A0A194QG26 A0A0K8TV62 A0A2W1BCF8 A0A194QHM5 A0A194RMF7 A0A2I6RJN8 J7FJ29 K7IVW0 A0A411K6Z1 A0A212ET38 A0A194QLW6 J7FJ25 A0A154PSU2 E9IRU5 A0A194QFW1 K7IVV9 A0A0K1YWI1 A0A232F1X5 D7EJS8
D5L0N2 A0A2Z6JJS6 A0A2A4K2A3 A0A2S1PSQ3 A0A2Z5CT64 A0A068F0U3 A0A2S1PSN1 A0A2Z5CT56 A0A2Z6JKL5 A0A1V0D9H5 A0A2Z5CT65 A0A2Z5CT45 A0A2Z6JKK7 A0A2Z5CTX4 A0A2Z6JQN4 A0A2Z6JSP9 A0A2Z5CVH7 A0A0K8TVQ8 A0A2Z6JKK5 A0A2Z6JIS5 A0A2Z6JIS9 A0A2Z6JJ49 A0A2Z6JVD1 A0A2Z5CVH9 A0A2Z5CT48 A0A2Z6JNK9 A0A2Z6JLZ3 A0A172MFA5 A0A2Z6JNK2 A0A2Z6JSR4 A0A2Z6JIS3 A0A2Z6JNJ2 A0A2Z6JIS1 A0A2Z5CV12 A0A2Z6JLY7 A0A1E1WI40 A0A2Z6JQP2 A0A2Z6JVD9 A0A2Z5CV54 A0A2Z5CTZ2 A0A2Z5CT49 A0A2Z5CTW1 A0A194RE58 A0A2Z6JJS2 A0A194PQ11 A0A2A4K2N2 D2JLJ8 A0A212FGZ1 A0A2Z5CU05 A0A2Z6JI71 A0A1V0D9G7 A0A0C5C563 A0A1V0D9G8 A0A2Z5CV07 A0A1V0D9G1 A0A1V0D9H1 A0A212ESQ5 A0A1V0D9G0 A0A068EU25 A0A3S2NRQ2 A0A194RM59 A0A194QLW1 A0A2W1BF80 A0A2H1V767 A0A2H1V738 A0A3G5EAS2 A0A2W1BMN3 A0A2A4JVD4 A0A0M9A0V9 A0A212FBK5 A0A286QUF5 A0A1W6L1S3 A0A248QJ37 A0A194QG26 A0A0K8TV62 A0A2W1BCF8 A0A194QHM5 A0A194RMF7 A0A2I6RJN8 J7FJ29 K7IVW0 A0A411K6Z1 A0A212ET38 A0A194QLW6 J7FJ25 A0A154PSU2 E9IRU5 A0A194QFW1 K7IVV9 A0A0K1YWI1 A0A232F1X5 D7EJS8
Pubmed
EMBL
EU435136
AB436837
AB436838
ABZ81072.1
BAM73792.1
BK010537
+ More
DAB41820.1 GU731538 ADE05588.1 MG583766 AXB26396.1 BK010514 DAB41797.1 KX443447 ASO98022.1 GU731537 ADE05587.1 BK010506 DAB41789.1 BK010504 NWSH01000248 DAB41787.1 PCG78034.1 MF278024 AWH61659.1 MG583775 AXB26405.1 KM016708 AID54860.1 MF278023 AWH61658.1 MG583772 AXB26402.1 BK010532 DAB41815.1 KY212077 ARA91641.1 MG583768 AXB26398.1 MG583762 AXB26392.1 BK010511 DAB41794.1 MG583774 AXB26404.1 BK010377 DAB41780.1 BK010376 DAB41779.1 MG583761 AXB26391.1 GCVX01000080 JAI18150.1 BK010501 DAB41784.1 BK010512 DAB41795.1 BK010533 DAB41816.1 BK010378 DAB41781.1 BK010374 DAB41777.1 MG583771 AXB26401.1 MG583767 AXB26397.1 BK010513 DAB41796.1 BK010510 DAB41793.1 KU145394 ANC96677.1 BK010503 DAB41786.1 BK010507 DAB41790.1 BK010502 DAB41785.1 BK010372 DAB41775.1 BK010371 DAB41774.1 MG583770 AXB26400.1 BK010500 DAB41783.1 BK010509 GDQN01004477 DAB41792.1 JAT86577.1 BK010508 DAB41791.1 BK010505 DAB41788.1 MG583769 AXB26399.1 MG583763 AXB26393.1 MG583765 AXB26395.1 MG583764 AXB26394.1 KQ460367 KPJ15565.1 BK010375 DAB41778.1 KQ459602 KPI93220.1 PCG78033.1 GQ915314 ACZ97408.3 AGBW02008584 OWR52994.1 MG583773 AXB26403.1 BK010373 DAB41776.1 KY212078 ARA91642.1 KP001135 AJN91180.1 KY212073 ARA91637.1 MG583760 AXB26390.1 KY212076 ARA91640.1 KY212072 ARA91636.1 AGBW02012725 OWR44536.1 KY212074 ARA91638.1 KM016707 AID54859.1 RSAL01000117 RVE46814.1 KQ459995 KPJ18519.1 KQ458981 KPJ04436.1 KZ150141 PZC72941.1 ODYU01001012 SOQ36636.1 SOQ36637.1 MH001164 AYW35337.1 PZC72943.1 NWSH01000502 PCG75997.1 KQ435774 KOX75070.1 AGBW02009303 OWR51116.1 KX425013 ASN63840.1 KX609539 ARN17946.1 KX443446 ASO98021.1 KPJ04442.1 GCVX01000057 JAI18173.1 PZC72942.1 KPJ04440.1 KPJ18515.1 KY304478 AUN86409.1 JX310101 AFP20612.1 MH500625 QBC73100.1 AGBW02012675 OWR44614.1 KPJ04441.1 JX310086 AFP20597.1 KQ435041 KZC14180.1 GL765283 EFZ16666.1 KPJ04438.1 KP859393 AKZ17703.1 NNAY01001296 OXU24448.1 KQ971342 EFA12854.2
DAB41820.1 GU731538 ADE05588.1 MG583766 AXB26396.1 BK010514 DAB41797.1 KX443447 ASO98022.1 GU731537 ADE05587.1 BK010506 DAB41789.1 BK010504 NWSH01000248 DAB41787.1 PCG78034.1 MF278024 AWH61659.1 MG583775 AXB26405.1 KM016708 AID54860.1 MF278023 AWH61658.1 MG583772 AXB26402.1 BK010532 DAB41815.1 KY212077 ARA91641.1 MG583768 AXB26398.1 MG583762 AXB26392.1 BK010511 DAB41794.1 MG583774 AXB26404.1 BK010377 DAB41780.1 BK010376 DAB41779.1 MG583761 AXB26391.1 GCVX01000080 JAI18150.1 BK010501 DAB41784.1 BK010512 DAB41795.1 BK010533 DAB41816.1 BK010378 DAB41781.1 BK010374 DAB41777.1 MG583771 AXB26401.1 MG583767 AXB26397.1 BK010513 DAB41796.1 BK010510 DAB41793.1 KU145394 ANC96677.1 BK010503 DAB41786.1 BK010507 DAB41790.1 BK010502 DAB41785.1 BK010372 DAB41775.1 BK010371 DAB41774.1 MG583770 AXB26400.1 BK010500 DAB41783.1 BK010509 GDQN01004477 DAB41792.1 JAT86577.1 BK010508 DAB41791.1 BK010505 DAB41788.1 MG583769 AXB26399.1 MG583763 AXB26393.1 MG583765 AXB26395.1 MG583764 AXB26394.1 KQ460367 KPJ15565.1 BK010375 DAB41778.1 KQ459602 KPI93220.1 PCG78033.1 GQ915314 ACZ97408.3 AGBW02008584 OWR52994.1 MG583773 AXB26403.1 BK010373 DAB41776.1 KY212078 ARA91642.1 KP001135 AJN91180.1 KY212073 ARA91637.1 MG583760 AXB26390.1 KY212076 ARA91640.1 KY212072 ARA91636.1 AGBW02012725 OWR44536.1 KY212074 ARA91638.1 KM016707 AID54859.1 RSAL01000117 RVE46814.1 KQ459995 KPJ18519.1 KQ458981 KPJ04436.1 KZ150141 PZC72941.1 ODYU01001012 SOQ36636.1 SOQ36637.1 MH001164 AYW35337.1 PZC72943.1 NWSH01000502 PCG75997.1 KQ435774 KOX75070.1 AGBW02009303 OWR51116.1 KX425013 ASN63840.1 KX609539 ARN17946.1 KX443446 ASO98021.1 KPJ04442.1 GCVX01000057 JAI18173.1 PZC72942.1 KPJ04440.1 KPJ18515.1 KY304478 AUN86409.1 JX310101 AFP20612.1 MH500625 QBC73100.1 AGBW02012675 OWR44614.1 KPJ04441.1 JX310086 AFP20597.1 KQ435041 KZC14180.1 GL765283 EFZ16666.1 KPJ04438.1 KP859393 AKZ17703.1 NNAY01001296 OXU24448.1 KQ971342 EFA12854.2
Proteomes
PRIDE
Pfam
PF00067 p450
Interpro
SUPFAM
SSF48264
SSF48264
Gene 3D
ProteinModelPortal
B1AAB5
A0A2Z6JJT5
D5L0N3
A0A2Z5CUB7
A0A2Z6JI91
A0A248QFE5
+ More
D5L0N2 A0A2Z6JJS6 A0A2A4K2A3 A0A2S1PSQ3 A0A2Z5CT64 A0A068F0U3 A0A2S1PSN1 A0A2Z5CT56 A0A2Z6JKL5 A0A1V0D9H5 A0A2Z5CT65 A0A2Z5CT45 A0A2Z6JKK7 A0A2Z5CTX4 A0A2Z6JQN4 A0A2Z6JSP9 A0A2Z5CVH7 A0A0K8TVQ8 A0A2Z6JKK5 A0A2Z6JIS5 A0A2Z6JIS9 A0A2Z6JJ49 A0A2Z6JVD1 A0A2Z5CVH9 A0A2Z5CT48 A0A2Z6JNK9 A0A2Z6JLZ3 A0A172MFA5 A0A2Z6JNK2 A0A2Z6JSR4 A0A2Z6JIS3 A0A2Z6JNJ2 A0A2Z6JIS1 A0A2Z5CV12 A0A2Z6JLY7 A0A1E1WI40 A0A2Z6JQP2 A0A2Z6JVD9 A0A2Z5CV54 A0A2Z5CTZ2 A0A2Z5CT49 A0A2Z5CTW1 A0A194RE58 A0A2Z6JJS2 A0A194PQ11 A0A2A4K2N2 D2JLJ8 A0A212FGZ1 A0A2Z5CU05 A0A2Z6JI71 A0A1V0D9G7 A0A0C5C563 A0A1V0D9G8 A0A2Z5CV07 A0A1V0D9G1 A0A1V0D9H1 A0A212ESQ5 A0A1V0D9G0 A0A068EU25 A0A3S2NRQ2 A0A194RM59 A0A194QLW1 A0A2W1BF80 A0A2H1V767 A0A2H1V738 A0A3G5EAS2 A0A2W1BMN3 A0A2A4JVD4 A0A0M9A0V9 A0A212FBK5 A0A286QUF5 A0A1W6L1S3 A0A248QJ37 A0A194QG26 A0A0K8TV62 A0A2W1BCF8 A0A194QHM5 A0A194RMF7 A0A2I6RJN8 J7FJ29 K7IVW0 A0A411K6Z1 A0A212ET38 A0A194QLW6 J7FJ25 A0A154PSU2 E9IRU5 A0A194QFW1 K7IVV9 A0A0K1YWI1 A0A232F1X5 D7EJS8
D5L0N2 A0A2Z6JJS6 A0A2A4K2A3 A0A2S1PSQ3 A0A2Z5CT64 A0A068F0U3 A0A2S1PSN1 A0A2Z5CT56 A0A2Z6JKL5 A0A1V0D9H5 A0A2Z5CT65 A0A2Z5CT45 A0A2Z6JKK7 A0A2Z5CTX4 A0A2Z6JQN4 A0A2Z6JSP9 A0A2Z5CVH7 A0A0K8TVQ8 A0A2Z6JKK5 A0A2Z6JIS5 A0A2Z6JIS9 A0A2Z6JJ49 A0A2Z6JVD1 A0A2Z5CVH9 A0A2Z5CT48 A0A2Z6JNK9 A0A2Z6JLZ3 A0A172MFA5 A0A2Z6JNK2 A0A2Z6JSR4 A0A2Z6JIS3 A0A2Z6JNJ2 A0A2Z6JIS1 A0A2Z5CV12 A0A2Z6JLY7 A0A1E1WI40 A0A2Z6JQP2 A0A2Z6JVD9 A0A2Z5CV54 A0A2Z5CTZ2 A0A2Z5CT49 A0A2Z5CTW1 A0A194RE58 A0A2Z6JJS2 A0A194PQ11 A0A2A4K2N2 D2JLJ8 A0A212FGZ1 A0A2Z5CU05 A0A2Z6JI71 A0A1V0D9G7 A0A0C5C563 A0A1V0D9G8 A0A2Z5CV07 A0A1V0D9G1 A0A1V0D9H1 A0A212ESQ5 A0A1V0D9G0 A0A068EU25 A0A3S2NRQ2 A0A194RM59 A0A194QLW1 A0A2W1BF80 A0A2H1V767 A0A2H1V738 A0A3G5EAS2 A0A2W1BMN3 A0A2A4JVD4 A0A0M9A0V9 A0A212FBK5 A0A286QUF5 A0A1W6L1S3 A0A248QJ37 A0A194QG26 A0A0K8TV62 A0A2W1BCF8 A0A194QHM5 A0A194RMF7 A0A2I6RJN8 J7FJ29 K7IVW0 A0A411K6Z1 A0A212ET38 A0A194QLW6 J7FJ25 A0A154PSU2 E9IRU5 A0A194QFW1 K7IVV9 A0A0K1YWI1 A0A232F1X5 D7EJS8
PDB
4D75
E-value=3.46408e-52,
Score=519
Ontologies
GO
Topology
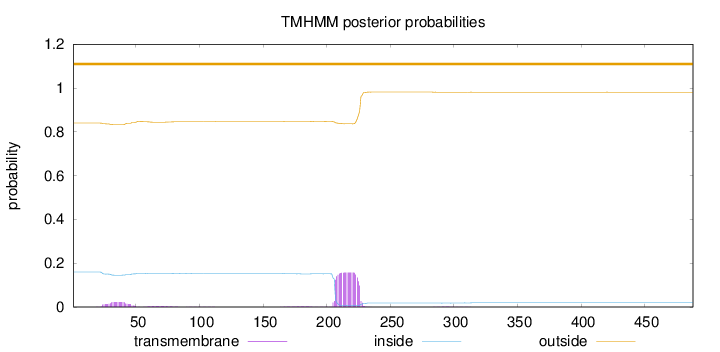
Length:
488
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.67273
Exp number, first 60 AAs:
0.44004
Total prob of N-in:
0.15987
outside
1 - 488
Population Genetic Test Statistics
Pi
290.93435
Theta
181.667406
Tajima's D
1.10355
CLR
1.279929
CSRT
0.685865706714664
Interpretation
Uncertain
