Gene
KWMTBOMO08852
Annotation
PREDICTED:_formin-binding_protein_1-like_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.042
Sequence
CDS
ATGAGTTGGGGTGTTGAGCTTTGGGATCAATATGATAACTTAGCCGCCCATACGCACAAAGGAATCGAGTTTTTAGATAAATATGGAAACTTTGTCAAGGAGAGGTGCGCTATAGAACTAGAATATGCAGGAAAACTCAGGAGGCTTGTCAAAAATTATCAACCAAAAAGGAAAGAAGAAGATGAATACCAGTATACAGCATGTAAAGCATTTAAACAGTTACTGCAAGAGTTAGGCGATTTCGCAGGACAACGGGAAGTTGTCGCGGAAAATCTTCAATCAAATGTAGTCCGAGAGTTGCACCTGCTCGCCAAAGAATTGCGAGAGGAAAGAAAGCAACATTTAAACGAAGGAGCAAAACAAATGGGAGTTCTGAACACTTCGATAGGTTCACTGGAACGAGCCAGGCGAGCGTACGAACGTGCGGCGAGGGAGTCGGAAAGAGCACTCGAGACCTTCCAAAAGGCAGACGCTGACCTCAATCTAAGTCGGGCAGAGTTGGAGAAGCAAAAAGCGAACATGAAACTCAGGAGTCAGGCGTGCGAAGATGCGAAACAGGAGTACCTAGAACAGCTTCGCAAAACGAACGAAGCCCAGAGGCAACACTACGAGCAGAGATTACCGCACGTGTTCAAACAGTTGCAAGACTTGGACGAGAAACGAATAAAGAATATTAAGAACTTTATGTTAAGTTCAGTGGATGTCGAAAGGAAAGTTTTTCCGATCATCATTCAGTGCTTTGACGGGATGGAACTGGCCGCAAATAGTATTAATGAAAAAGAGGACACAAAATTAGTTATAGACAGGTACAAGTCTGGTTTCGTTCCGCCCGAAGATTTCCAGTTCGAGTTGGCTTCCGGGGCAGACACAACAGACTCGGCGTCGACCCATCACCAGCCGCACAACCACCTGACGGCGTCCCGGGGGACCGTCTCCGGGAACAAGCTAAAGAAACGCGGCGGTCTACTCTCCATATTCAGTTCGAATAAGAACAACATGTCTACGGATGGAAAGGAGGACTATTCAGATTTGCCACCGAATCAAAAGAAAAAGAAACTTCAAGCTAAAGTACAAGAACTCAGTAAACAGGTTGCGCAAGAACAAGCCGCAATGGAAGGTCTTATGAAAATGAAAGGAGTGTACGAAATGAGCCCCGCGTTGGGTGATCCAATGACTGTAGAAGGTCAACTTAACGAATGCTGCGATAAATTGAAGAAACTTCGCTCGCAACTGAACAAGTTCGAGGAGCTCTTAGCCGAAGCGAACAGTCAAGTGAACGCGGCTAACTCGTTACACCCCGTCGCCAAGACTAATGGCGCGCCGCCCACGCAGGCTACAAGTATCGGTTCGAACAGCGAGTCCCTGTCCCGCTCGGCGTCGGAGTCGTCTGTGAGTACGGGCACGGGCACGGGCGGAGGGGCGGGGGTGCGCGCGGCGGGAGGCTCGCCCGAGTCCGGCCTGGGCGGGGAGCTCGCCGCCGCACACACAGACCACGCCAACGGCGAGCACGACCAGGACGACCGCGAGTCCGACCTCGACTACTACTACTGCGAGCCCGACCTGCAACCCGTCGGCTACTGCAAGGCGCTTTATGCCTTCGAAGGTATCGGCAGCGGGTCCACGATGCGGATGGAGTGCGGCGAGCAGCTGCTGGTGCTGGAGACGGACGCGGGGGACGGCTGGACGCGCGTGCGCCGCAACCGCACCCGCGAGGAGGGGTTCGTGCCCACAACGTACATCGCGACCACGCTCTACTCCGACGCGCAACACTAG
Protein
MSWGVELWDQYDNLAAHTHKGIEFLDKYGNFVKERCAIELEYAGKLRRLVKNYQPKRKEEDEYQYTACKAFKQLLQELGDFAGQREVVAENLQSNVVRELHLLAKELREERKQHLNEGAKQMGVLNTSIGSLERARRAYERAARESERALETFQKADADLNLSRAELEKQKANMKLRSQACEDAKQEYLEQLRKTNEAQRQHYEQRLPHVFKQLQDLDEKRIKNIKNFMLSSVDVERKVFPIIIQCFDGMELAANSINEKEDTKLVIDRYKSGFVPPEDFQFELASGADTTDSASTHHQPHNHLTASRGTVSGNKLKKRGGLLSIFSSNKNNMSTDGKEDYSDLPPNQKKKKLQAKVQELSKQVAQEQAAMEGLMKMKGVYEMSPALGDPMTVEGQLNECCDKLKKLRSQLNKFEELLAEANSQVNAANSLHPVAKTNGAPPTQATSIGSNSESLSRSASESSVSTGTGTGGGAGVRAAGGSPESGLGGELAAAHTDHANGEHDQDDRESDLDYYYCEPDLQPVGYCKALYAFEGIGSGSTMRMECGEQLLVLETDAGDGWTRVRRNRTREEGFVPTTYIATTLYSDAQH
Summary
Uniprot
A0A2A4J3R1
A0A2A4J4X0
A0A437B8A1
A0A212F4C0
A0A1E1WI75
A0A194RCK6
+ More
A0A194PWQ5 A0A2H1WJ05 A0A0C9R089 A0A0C9RCH0 A0A1B6K8J1 A0A0K8TM08 A0A1B6HL70 A0A023F3U4 A0A195BPZ7 A0A195CEZ2 X1WJA7 A0A151JN84 A0A1Q3F9J4 A0A1Q3F9E0 A0A1B6FEE4 A0A1B6J3R0 E2A0X3 A0A1L8E5L6 A0A1Q3F998 A0A158NV61 A0A1Q3F9G9 B0XBC5 A0A1B6CBX1 A0A1I8Q6U8 A0A1B6C690 V9ID65 A0A1L8E596 T1PDW1 A0A195F3T7 V9IB00 A0A1I8Q6S9 A0A139WB00 A0A0V0GAK2 K7IRH3 A0A1W4WWA2 A0A1W4X743 A0A154PJW8 A0A139WB46 A0A3L8DMN7 M9PE52 M9NE16 A0A1B6KPT7 A0A1B6K8T0 A0A0M9A9R0 A0A0J9RQD6 A0A0J9UEG8 M9ND44 A0A0P8XT52 A0A0P8YFQ9 A0A1B6KPI6 A0A0Q5UI58 A0A0Q5U3Z9 A0A088ACC6 A0A0P9C273 A0A0P8ZTA7 A0A0R1E0Y0 A0A0R1DVH5 A0A2A3EHY4 A0A1B6JWX0 A0A3B0JC37 A0A3B0JQ77 E2C6T0 A0A0A9VVX5 A0A0A9VUG6 A0A1A9ZBZ7 A0A1A9XQW2 A0A1B0ANS1 A0A1A9UP53 A0A0Q9XML2 M9NF07 M9NF10 A0A0J9RNY1 A0A0J9UEH9 A0A0Q5U3A7 A0A0Q5U4C3 M9PBP3 A0A0R1DVS8 A0A0R1DVN2 A0A1B6KUJ5 A0A1S3DBA0 A0A1B6KDB0 A0A0J9RPE4 A0A0R3P427 Q2M010 A0A0P8Y8K5 A0A1W4X5Q7 A0A1B6KCE3 A0A1Y1LZV8 A0A336MFA7 A0A1B6LXW5 A0A0Q5UA47 A0A0R1DXF7 A0A1Y1LYK7 A0A0K8SEY7
A0A194PWQ5 A0A2H1WJ05 A0A0C9R089 A0A0C9RCH0 A0A1B6K8J1 A0A0K8TM08 A0A1B6HL70 A0A023F3U4 A0A195BPZ7 A0A195CEZ2 X1WJA7 A0A151JN84 A0A1Q3F9J4 A0A1Q3F9E0 A0A1B6FEE4 A0A1B6J3R0 E2A0X3 A0A1L8E5L6 A0A1Q3F998 A0A158NV61 A0A1Q3F9G9 B0XBC5 A0A1B6CBX1 A0A1I8Q6U8 A0A1B6C690 V9ID65 A0A1L8E596 T1PDW1 A0A195F3T7 V9IB00 A0A1I8Q6S9 A0A139WB00 A0A0V0GAK2 K7IRH3 A0A1W4WWA2 A0A1W4X743 A0A154PJW8 A0A139WB46 A0A3L8DMN7 M9PE52 M9NE16 A0A1B6KPT7 A0A1B6K8T0 A0A0M9A9R0 A0A0J9RQD6 A0A0J9UEG8 M9ND44 A0A0P8XT52 A0A0P8YFQ9 A0A1B6KPI6 A0A0Q5UI58 A0A0Q5U3Z9 A0A088ACC6 A0A0P9C273 A0A0P8ZTA7 A0A0R1E0Y0 A0A0R1DVH5 A0A2A3EHY4 A0A1B6JWX0 A0A3B0JC37 A0A3B0JQ77 E2C6T0 A0A0A9VVX5 A0A0A9VUG6 A0A1A9ZBZ7 A0A1A9XQW2 A0A1B0ANS1 A0A1A9UP53 A0A0Q9XML2 M9NF07 M9NF10 A0A0J9RNY1 A0A0J9UEH9 A0A0Q5U3A7 A0A0Q5U4C3 M9PBP3 A0A0R1DVS8 A0A0R1DVN2 A0A1B6KUJ5 A0A1S3DBA0 A0A1B6KDB0 A0A0J9RPE4 A0A0R3P427 Q2M010 A0A0P8Y8K5 A0A1W4X5Q7 A0A1B6KCE3 A0A1Y1LZV8 A0A336MFA7 A0A1B6LXW5 A0A0Q5UA47 A0A0R1DXF7 A0A1Y1LYK7 A0A0K8SEY7
Pubmed
EMBL
NWSH01003204
PCG66787.1
PCG66786.1
RSAL01000127
RVE46529.1
AGBW02010386
+ More
OWR48586.1 GDQN01004344 JAT86710.1 KQ460367 KPJ15568.1 KQ459596 KPI95565.1 ODYU01008981 SOQ53038.1 GBYB01001385 JAG71152.1 GBYB01010782 JAG80549.1 GEBQ01032211 GEBQ01008488 JAT07766.1 JAT31489.1 GDAI01002201 JAI15402.1 GECU01032264 JAS75442.1 GBBI01002641 JAC16071.1 KQ976424 KYM88208.1 KQ977827 KYM99459.1 ABLF02022819 ABLF02022824 ABLF02022825 KQ978850 KYN27956.1 GFDL01010856 JAV24189.1 GFDL01010893 JAV24152.1 GECZ01021201 JAS48568.1 GECU01013888 JAS93818.1 GL435626 EFN73026.1 GFDF01000165 JAV13919.1 GFDL01010891 JAV24154.1 ADTU01026907 ADTU01026908 GFDL01010835 JAV24210.1 DS232624 EDS44216.1 GEDC01026351 GEDC01015813 JAS10947.1 JAS21485.1 GEDC01028374 GEDC01008590 JAS08924.1 JAS28708.1 JR038465 AEY58264.1 GFDF01000164 JAV13920.1 KA646123 AFP60752.1 KQ981820 KYN35245.1 JR038466 AEY58265.1 KQ971377 KYB25114.1 GECL01000921 JAP05203.1 KQ434923 KZC11510.1 KYB25113.1 QOIP01000006 RLU21677.1 AE014296 AGB94108.1 AFH04290.1 AFH04291.1 GEBQ01026743 JAT13234.1 GEBQ01032124 JAT07853.1 KQ435712 KOX79376.1 CM002912 KMY97579.1 KMY97575.1 AFH04292.1 CH902618 KPU77765.1 KPU77766.1 GEBQ01028760 GEBQ01026843 JAT11217.1 JAT13134.1 CH954178 KQS43578.1 KQS43579.1 KPU77767.1 KPU77768.1 CM000159 KRK01162.1 KRK01163.1 KZ288256 PBC30621.1 GECU01003998 JAT03709.1 OUUW01000002 SPP77622.1 SPP77620.1 GL453161 EFN76392.1 GBHO01044253 GBRD01013975 JAF99350.1 JAG51851.1 GBHO01044255 GBRD01013976 JAF99348.1 JAG51850.1 JXJN01000938 CH933809 KRG06158.1 AFH04288.1 AFH04293.1 KMY97578.1 KMY97585.1 KQS43583.1 KQS43577.1 AGB94107.1 KRK01161.1 KRK01164.1 GEBQ01024874 JAT15103.1 GEBQ01030560 JAT09417.1 KMY97582.1 CH379069 KRT07881.1 EAL31127.3 KPU77769.1 GEBQ01030871 GEBQ01004356 JAT09106.1 JAT35621.1 GEZM01043963 JAV78631.1 UFQT01001123 SSX29034.1 GEBQ01021225 GEBQ01011431 JAT18752.1 JAT28546.1 KQS43581.1 KRK01166.1 GEZM01043962 JAV78632.1 GBRD01013974 JAG51852.1
OWR48586.1 GDQN01004344 JAT86710.1 KQ460367 KPJ15568.1 KQ459596 KPI95565.1 ODYU01008981 SOQ53038.1 GBYB01001385 JAG71152.1 GBYB01010782 JAG80549.1 GEBQ01032211 GEBQ01008488 JAT07766.1 JAT31489.1 GDAI01002201 JAI15402.1 GECU01032264 JAS75442.1 GBBI01002641 JAC16071.1 KQ976424 KYM88208.1 KQ977827 KYM99459.1 ABLF02022819 ABLF02022824 ABLF02022825 KQ978850 KYN27956.1 GFDL01010856 JAV24189.1 GFDL01010893 JAV24152.1 GECZ01021201 JAS48568.1 GECU01013888 JAS93818.1 GL435626 EFN73026.1 GFDF01000165 JAV13919.1 GFDL01010891 JAV24154.1 ADTU01026907 ADTU01026908 GFDL01010835 JAV24210.1 DS232624 EDS44216.1 GEDC01026351 GEDC01015813 JAS10947.1 JAS21485.1 GEDC01028374 GEDC01008590 JAS08924.1 JAS28708.1 JR038465 AEY58264.1 GFDF01000164 JAV13920.1 KA646123 AFP60752.1 KQ981820 KYN35245.1 JR038466 AEY58265.1 KQ971377 KYB25114.1 GECL01000921 JAP05203.1 KQ434923 KZC11510.1 KYB25113.1 QOIP01000006 RLU21677.1 AE014296 AGB94108.1 AFH04290.1 AFH04291.1 GEBQ01026743 JAT13234.1 GEBQ01032124 JAT07853.1 KQ435712 KOX79376.1 CM002912 KMY97579.1 KMY97575.1 AFH04292.1 CH902618 KPU77765.1 KPU77766.1 GEBQ01028760 GEBQ01026843 JAT11217.1 JAT13134.1 CH954178 KQS43578.1 KQS43579.1 KPU77767.1 KPU77768.1 CM000159 KRK01162.1 KRK01163.1 KZ288256 PBC30621.1 GECU01003998 JAT03709.1 OUUW01000002 SPP77622.1 SPP77620.1 GL453161 EFN76392.1 GBHO01044253 GBRD01013975 JAF99350.1 JAG51851.1 GBHO01044255 GBRD01013976 JAF99348.1 JAG51850.1 JXJN01000938 CH933809 KRG06158.1 AFH04288.1 AFH04293.1 KMY97578.1 KMY97585.1 KQS43583.1 KQS43577.1 AGB94107.1 KRK01161.1 KRK01164.1 GEBQ01024874 JAT15103.1 GEBQ01030560 JAT09417.1 KMY97582.1 CH379069 KRT07881.1 EAL31127.3 KPU77769.1 GEBQ01030871 GEBQ01004356 JAT09106.1 JAT35621.1 GEZM01043963 JAV78631.1 UFQT01001123 SSX29034.1 GEBQ01021225 GEBQ01011431 JAT18752.1 JAT28546.1 KQS43581.1 KRK01166.1 GEZM01043962 JAV78632.1 GBRD01013974 JAG51852.1
Proteomes
UP000218220
UP000283053
UP000007151
UP000053240
UP000053268
UP000078540
+ More
UP000078542 UP000007819 UP000078492 UP000000311 UP000005205 UP000002320 UP000095300 UP000095301 UP000078541 UP000007266 UP000002358 UP000192223 UP000076502 UP000279307 UP000000803 UP000053105 UP000007801 UP000008711 UP000005203 UP000002282 UP000242457 UP000268350 UP000008237 UP000092445 UP000092443 UP000092460 UP000078200 UP000009192 UP000079169 UP000001819
UP000078542 UP000007819 UP000078492 UP000000311 UP000005205 UP000002320 UP000095300 UP000095301 UP000078541 UP000007266 UP000002358 UP000192223 UP000076502 UP000279307 UP000000803 UP000053105 UP000007801 UP000008711 UP000005203 UP000002282 UP000242457 UP000268350 UP000008237 UP000092445 UP000092443 UP000092460 UP000078200 UP000009192 UP000079169 UP000001819
Interpro
Gene 3D
ProteinModelPortal
A0A2A4J3R1
A0A2A4J4X0
A0A437B8A1
A0A212F4C0
A0A1E1WI75
A0A194RCK6
+ More
A0A194PWQ5 A0A2H1WJ05 A0A0C9R089 A0A0C9RCH0 A0A1B6K8J1 A0A0K8TM08 A0A1B6HL70 A0A023F3U4 A0A195BPZ7 A0A195CEZ2 X1WJA7 A0A151JN84 A0A1Q3F9J4 A0A1Q3F9E0 A0A1B6FEE4 A0A1B6J3R0 E2A0X3 A0A1L8E5L6 A0A1Q3F998 A0A158NV61 A0A1Q3F9G9 B0XBC5 A0A1B6CBX1 A0A1I8Q6U8 A0A1B6C690 V9ID65 A0A1L8E596 T1PDW1 A0A195F3T7 V9IB00 A0A1I8Q6S9 A0A139WB00 A0A0V0GAK2 K7IRH3 A0A1W4WWA2 A0A1W4X743 A0A154PJW8 A0A139WB46 A0A3L8DMN7 M9PE52 M9NE16 A0A1B6KPT7 A0A1B6K8T0 A0A0M9A9R0 A0A0J9RQD6 A0A0J9UEG8 M9ND44 A0A0P8XT52 A0A0P8YFQ9 A0A1B6KPI6 A0A0Q5UI58 A0A0Q5U3Z9 A0A088ACC6 A0A0P9C273 A0A0P8ZTA7 A0A0R1E0Y0 A0A0R1DVH5 A0A2A3EHY4 A0A1B6JWX0 A0A3B0JC37 A0A3B0JQ77 E2C6T0 A0A0A9VVX5 A0A0A9VUG6 A0A1A9ZBZ7 A0A1A9XQW2 A0A1B0ANS1 A0A1A9UP53 A0A0Q9XML2 M9NF07 M9NF10 A0A0J9RNY1 A0A0J9UEH9 A0A0Q5U3A7 A0A0Q5U4C3 M9PBP3 A0A0R1DVS8 A0A0R1DVN2 A0A1B6KUJ5 A0A1S3DBA0 A0A1B6KDB0 A0A0J9RPE4 A0A0R3P427 Q2M010 A0A0P8Y8K5 A0A1W4X5Q7 A0A1B6KCE3 A0A1Y1LZV8 A0A336MFA7 A0A1B6LXW5 A0A0Q5UA47 A0A0R1DXF7 A0A1Y1LYK7 A0A0K8SEY7
A0A194PWQ5 A0A2H1WJ05 A0A0C9R089 A0A0C9RCH0 A0A1B6K8J1 A0A0K8TM08 A0A1B6HL70 A0A023F3U4 A0A195BPZ7 A0A195CEZ2 X1WJA7 A0A151JN84 A0A1Q3F9J4 A0A1Q3F9E0 A0A1B6FEE4 A0A1B6J3R0 E2A0X3 A0A1L8E5L6 A0A1Q3F998 A0A158NV61 A0A1Q3F9G9 B0XBC5 A0A1B6CBX1 A0A1I8Q6U8 A0A1B6C690 V9ID65 A0A1L8E596 T1PDW1 A0A195F3T7 V9IB00 A0A1I8Q6S9 A0A139WB00 A0A0V0GAK2 K7IRH3 A0A1W4WWA2 A0A1W4X743 A0A154PJW8 A0A139WB46 A0A3L8DMN7 M9PE52 M9NE16 A0A1B6KPT7 A0A1B6K8T0 A0A0M9A9R0 A0A0J9RQD6 A0A0J9UEG8 M9ND44 A0A0P8XT52 A0A0P8YFQ9 A0A1B6KPI6 A0A0Q5UI58 A0A0Q5U3Z9 A0A088ACC6 A0A0P9C273 A0A0P8ZTA7 A0A0R1E0Y0 A0A0R1DVH5 A0A2A3EHY4 A0A1B6JWX0 A0A3B0JC37 A0A3B0JQ77 E2C6T0 A0A0A9VVX5 A0A0A9VUG6 A0A1A9ZBZ7 A0A1A9XQW2 A0A1B0ANS1 A0A1A9UP53 A0A0Q9XML2 M9NF07 M9NF10 A0A0J9RNY1 A0A0J9UEH9 A0A0Q5U3A7 A0A0Q5U4C3 M9PBP3 A0A0R1DVS8 A0A0R1DVN2 A0A1B6KUJ5 A0A1S3DBA0 A0A1B6KDB0 A0A0J9RPE4 A0A0R3P427 Q2M010 A0A0P8Y8K5 A0A1W4X5Q7 A0A1B6KCE3 A0A1Y1LZV8 A0A336MFA7 A0A1B6LXW5 A0A0Q5UA47 A0A0R1DXF7 A0A1Y1LYK7 A0A0K8SEY7
PDB
2EFL
E-value=2.10418e-60,
Score=591
Ontologies
GO
GO:0016021
GO:0005543
GO:0007010
GO:0005886
GO:0097320
GO:0008092
GO:0005737
GO:0005856
GO:0031594
GO:0007009
GO:0008582
GO:0051126
GO:0035317
GO:0008289
GO:0032794
GO:0034613
GO:0007349
GO:0030708
GO:0007498
GO:0008104
GO:0045886
GO:1903391
GO:0030496
GO:0032154
GO:0012506
GO:0030837
GO:0005515
GO:0016311
GO:0016791
GO:0008270
GO:0015074
GO:0016817
GO:0043565
GO:0016876
GO:0043039
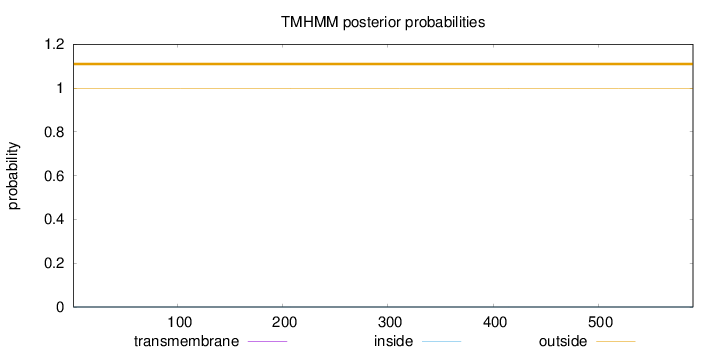
Topology
Length:
590
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00016
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00040
outside
1 - 590
Population Genetic Test Statistics
Pi
234.234684
Theta
186.270955
Tajima's D
0.815626
CLR
0.329333
CSRT
0.601669916504175
Interpretation
Uncertain
