Gene
KWMTBOMO08850
Pre Gene Modal
BGIBMGA007847
Annotation
PREDICTED:_myotubularin-related_protein_2_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.31 Nuclear Reliability : 1.551
Sequence
CDS
ATGGATAAACATAACAGTTCAGAACTATTAAGTTCCGACTTTTCGCTATCCAAGAATGCAAGTTCTGATTCATTGGATTCTGATTCTAAATCAAGTTCTTTGAATTCTAAACACGGACAGGACTCCCCCCATGTCCTCGGAGATATACAGCTGTTGGATGGTGAAAAGGTTACAAGTGTAGCTCGTGATGTAACTTATTTGTGCCCATACAGTGGACCAACGAGGGGAGTCCTGAAAGTGACAAACTATCAACTTCACTTTAGACCAACTGATCCTAGTGCTACTCTAGGTACATTGAGTGTGCCACTGGGGGTTGTTTCTCGAATTGAAAAAGTTGGTGGTGCTTCTTCCAAAGGTGAAAACTCATATGGAATAGAAATTTTTTGTAAGGATATGAGGAACTTAAGATTTGCACACAGACAAGAGAATCATTCACGTCGTGGTATATTTGAAAAACTGCAGCAGCTAGCTTTTCCTCTGTCACACCGACTTCCGATGTTTGCATTCAGTTATTCTGAAAGTTTCCCTGAAGATGGTTGGCATGTGTATGAACCCATTGCTGAGCTGAGACGAATGGGTGTAAGCAATGACATGTGGCGCATAACACGAATAAACGATAAGTATGAAGTGTGTGATAGCTATCCGTCTATATGGGCAGTCCCTTCTGCAGCTGATGATGATTTACTACGATCAGTGGCAACATTTAGATCACGTGGCCGCATTCCTGTACTATCCTGGATTCATCCTAACTCCCAAGCTACAATAACCAGGTGCAGTCAACCGTTAGTTGGGGTAAGCGGCAAGCGAAGTCGTGAAGATGAAAGGTACATTCAACTAATTATGGATGCAAATGCCCAGGCTCATAAACTTTTCATCATGGATGCTCGTCCCACTGCTAATGCTGTCGCCAATAAAGCCAAAGGAGGAGGTTACGAGTCTGAAGACGCGTATCAGAATGCGGATCTGGTGTTCTTGGACATTCATAACATTCACGTCATGAGAGAAAGCTTACGGAAACTAAAGGAATTATGCTTTCCACAGATTGATCAATCCAAATGGTTCAGCGGAATAGAAGCCAGCTGTTGGCTGAAGCACATCAAATGTATTCTGGCTGGTGCTGTCCGTATTGTTGATAAGGTTGAGAATCATAAGACGTCGGTGCTCGTGCATTGTTCGGATGGCTGGGACCGCACGGCTCAACTGACGGCGCTGTCGATGTTAATGCTCGACCCTTACTACAGAACACTTCGAGGATTCCAAGTTCTAATAGAAAAGGAGTGGTTATCTTTTGGACATAAATTTCAATTGCGCATCGGGCACGGCGACGAGCGACACTCGGACGCGGACCGCTCGCCCGTGTTCGTGCAGTGGGTGGACTGCGTGTGGCAGCTGCAGCAGCAGTTCCCGACGGCGTTCGAATTCACAGAACGACTCCTAATAACTATAGTCGATCACCTATATTCGTGTCGCTTCGGAACCTTCCTGTTCAATACTGAACGGGAAAGGGTAAAGGAAGAAGTGAAGACTAAGACCGTATCTCTATGGTCTTATATAAATAGTCGTCAAGATTTGTATTTGAATGCGCTTTACTGGGGCTCGTCGTCCGGCGCCACGTCGCCCTCCTCGCCGTACTCGAGGCCTCCCGTCGTGCTAGTTCCTGTCGCTTCATTAAGGGTCATCAAACTGTGGAAAGCGCTGTACTGCAGATGGAACCCAATAATGAGGCAGCAGGATCCGATATACCAAAGATCACGTGAGCTGGTGGCTCTCCGCGCGCAGCTGGAGGCGGCGGTGGCGGCGGCCGCCGGGGACCACGCCGCCAAGCAACACCGCCGCGGCCTCACGCCGCCCAGGGTCGCCAGCCCGCACCATGTCTGA
Protein
MDKHNSSELLSSDFSLSKNASSDSLDSDSKSSSLNSKHGQDSPHVLGDIQLLDGEKVTSVARDVTYLCPYSGPTRGVLKVTNYQLHFRPTDPSATLGTLSVPLGVVSRIEKVGGASSKGENSYGIEIFCKDMRNLRFAHRQENHSRRGIFEKLQQLAFPLSHRLPMFAFSYSESFPEDGWHVYEPIAELRRMGVSNDMWRITRINDKYEVCDSYPSIWAVPSAADDDLLRSVATFRSRGRIPVLSWIHPNSQATITRCSQPLVGVSGKRSREDERYIQLIMDANAQAHKLFIMDARPTANAVANKAKGGGYESEDAYQNADLVFLDIHNIHVMRESLRKLKELCFPQIDQSKWFSGIEASCWLKHIKCILAGAVRIVDKVENHKTSVLVHCSDGWDRTAQLTALSMLMLDPYYRTLRGFQVLIEKEWLSFGHKFQLRIGHGDERHSDADRSPVFVQWVDCVWQLQQQFPTAFEFTERLLITIVDHLYSCRFGTFLFNTERERVKEEVKTKTVSLWSYINSRQDLYLNALYWGSSSGATSPSSPYSRPPVVLVPVASLRVIKLWKALYCRWNPIMRQQDPIYQRSRELVALRAQLEAAVAAAAGDHAAKQHRRGLTPPRVASPHHV
Summary
Uniprot
A0A2H1WKH1
A0A2A4JBE7
A0A194RDN7
A0A437B033
A0A2J7Q1C2
A0A195D504
+ More
A0A067R4P3 A0A026WLL8 A0A3L8E7Q5 A0A1B6MD37 A0A1B6L6A7 D6W9S3 A0A0C9QN08 A0A0C9R1G5 A0A0C9R1G2 A0A1W4X484 A0A195DJ47 A0A195BQW0 K7ITS3 A0A158NQM9 A0A232F5U2 A0A195ERR0 T1I1D3 A0A0T6ATR8 E1ZXD1 A0A151XB23 E2B9S6 A0A1B6CJN7 A0A1B6CSM6 A0A088ACG1 A0A2A3EFN7 A0A1Y1KRF7 A0A0P4VRX4 A0A310STE7 A0A0L7R5C2 N6SXW9 J3JYW5 A0A1B6FUB7 A0A069DVU4 A0A1B6K0N9 A0A0C9RNB0 A0A1B6HZS2 A0A1S3CXK2 A0A2S2R6L7 A0A0V0G8M7 A0A2H8TCN2 J9JNR1 A0A0M9A8G1 A0A154PHY4 A0A0J7P0V1 E0VHL9 S4P7Q9 A0A1B0D1E8 A0A0A9Y770 A0A1L8DYY1 T1IY29 A0A182H1G5 A0A0P6J551 A0A0P5VTG8 A0A0N8AGG2 A0A2M4AHL3 A0A1Q3G0W1 A0A182JVD7 A0A2M4AHG4 A0A0P4ZNA5 A0A182QW07 A0A182VV39 A0A182R9C9 A0A182NLJ6 A0A182JG63 A0A336K5U4 A0A1S4H1F2 A0A182PB34 A0A182VBA3 A0A182TZ48 A0A182XCL7 A0A182L5V7 A0A182IDZ3 Q7Q1Z5 A0A084W3R2 A0A182F216 W5J5L9 A0A0P6HGU3 A0A1I8M937 A0A1B6EZR2 R7UQ41 A0A182M6M2 T1PJL1 A0A0L0CA33 A0A034V144 A0A0K8UXI6 A0A1A9YDW5 A0A1B0BZJ2 A0A1A9UNK3 A0A1B0A5M0 W8BSE7 A0A0N8E1T3 A0A1J1HER9 A0A1B0FDL8 A0A0A1X3Q1
A0A067R4P3 A0A026WLL8 A0A3L8E7Q5 A0A1B6MD37 A0A1B6L6A7 D6W9S3 A0A0C9QN08 A0A0C9R1G5 A0A0C9R1G2 A0A1W4X484 A0A195DJ47 A0A195BQW0 K7ITS3 A0A158NQM9 A0A232F5U2 A0A195ERR0 T1I1D3 A0A0T6ATR8 E1ZXD1 A0A151XB23 E2B9S6 A0A1B6CJN7 A0A1B6CSM6 A0A088ACG1 A0A2A3EFN7 A0A1Y1KRF7 A0A0P4VRX4 A0A310STE7 A0A0L7R5C2 N6SXW9 J3JYW5 A0A1B6FUB7 A0A069DVU4 A0A1B6K0N9 A0A0C9RNB0 A0A1B6HZS2 A0A1S3CXK2 A0A2S2R6L7 A0A0V0G8M7 A0A2H8TCN2 J9JNR1 A0A0M9A8G1 A0A154PHY4 A0A0J7P0V1 E0VHL9 S4P7Q9 A0A1B0D1E8 A0A0A9Y770 A0A1L8DYY1 T1IY29 A0A182H1G5 A0A0P6J551 A0A0P5VTG8 A0A0N8AGG2 A0A2M4AHL3 A0A1Q3G0W1 A0A182JVD7 A0A2M4AHG4 A0A0P4ZNA5 A0A182QW07 A0A182VV39 A0A182R9C9 A0A182NLJ6 A0A182JG63 A0A336K5U4 A0A1S4H1F2 A0A182PB34 A0A182VBA3 A0A182TZ48 A0A182XCL7 A0A182L5V7 A0A182IDZ3 Q7Q1Z5 A0A084W3R2 A0A182F216 W5J5L9 A0A0P6HGU3 A0A1I8M937 A0A1B6EZR2 R7UQ41 A0A182M6M2 T1PJL1 A0A0L0CA33 A0A034V144 A0A0K8UXI6 A0A1A9YDW5 A0A1B0BZJ2 A0A1A9UNK3 A0A1B0A5M0 W8BSE7 A0A0N8E1T3 A0A1J1HER9 A0A1B0FDL8 A0A0A1X3Q1
Pubmed
EMBL
ODYU01008934
SOQ52954.1
NWSH01002160
PCG69018.1
KQ460367
KPJ15569.1
+ More
RSAL01000249 RVE43495.1 NEVH01019421 PNF22378.1 KQ976885 KYN07509.1 KK852703 KDR18081.1 KK107154 EZA56848.1 QOIP01000001 RLU27558.1 GEBQ01006141 JAT33836.1 GEBQ01020750 JAT19227.1 KQ971312 EEZ98113.2 GBYB01002042 JAG71809.1 GBYB01001790 GBYB01002043 JAG71557.1 JAG71810.1 GBYB01001780 JAG71547.1 KQ980824 KYN12524.1 KQ976419 KYM89268.1 ADTU01023417 NNAY01000927 OXU25860.1 KQ982021 KYN30587.1 ACPB03005173 LJIG01022876 KRT78277.1 GL435030 EFN74221.1 KQ982335 KYQ57547.1 GL446605 EFN87513.1 GEDC01029504 GEDC01023686 JAS07794.1 JAS13612.1 GEDC01020921 GEDC01009377 JAS16377.1 JAS27921.1 KZ288256 PBC30603.1 GEZM01078343 JAV62760.1 GDKW01001265 JAI55330.1 KQ760132 OAD61801.1 KQ414654 KOC66019.1 APGK01052521 APGK01052522 KB741213 ENN72599.1 BT128446 AEE63403.1 GECZ01015952 JAS53817.1 GBGD01000859 JAC88030.1 GECU01026144 GECU01002689 JAS81562.1 JAT05018.1 GBYB01014977 JAG84744.1 GECU01027543 GECU01014113 JAS80163.1 JAS93593.1 GGMS01016473 MBY85676.1 GECL01001806 JAP04318.1 GFXV01000071 MBW11876.1 ABLF02039071 KQ435712 KOX79357.1 KQ434923 KZC11486.1 LBMM01000464 KMQ98275.1 DS235171 EEB12905.1 GAIX01004484 JAA88076.1 AJVK01010231 GBHO01016137 GBHO01016136 GBHO01016133 GBHO01005438 GBRD01008408 JAG27467.1 JAG27468.1 JAG27471.1 JAG38166.1 JAG57413.1 GFDF01002404 JAV11680.1 JH431669 JXUM01103444 KQ564805 KXJ71726.1 GDUN01000683 JAN95236.1 GDIP01095846 JAM07869.1 GDIP01149841 GDIQ01035029 LRGB01000005 JAJ73561.1 JAN59708.1 KZS22012.1 GGFK01006964 MBW40285.1 GFDL01001615 JAV33430.1 GGFK01006899 MBW40220.1 GDIP01214080 JAJ09322.1 AXCN02002502 UFQS01000093 UFQT01000093 SSW99222.1 SSX19602.1 AAAB01008839 AAAB01008980 APCN01004541 EAA14432.3 ATLV01020113 KE525294 KFB44856.1 ADMH02002162 ETN58165.1 GDIQ01019032 JAN75705.1 GECZ01026257 JAS43512.1 AMQN01006822 KB299236 ELU08223.1 AXCM01004598 KA648128 AFP62757.1 JRES01000699 KNC29075.1 GAKP01023438 JAC35520.1 GDHF01021234 JAI31080.1 JXJN01023173 GAMC01006727 JAB99828.1 GDIQ01070204 JAN24533.1 CVRI01000001 CRK86447.1 CCAG010007366 GBXI01008555 JAD05737.1
RSAL01000249 RVE43495.1 NEVH01019421 PNF22378.1 KQ976885 KYN07509.1 KK852703 KDR18081.1 KK107154 EZA56848.1 QOIP01000001 RLU27558.1 GEBQ01006141 JAT33836.1 GEBQ01020750 JAT19227.1 KQ971312 EEZ98113.2 GBYB01002042 JAG71809.1 GBYB01001790 GBYB01002043 JAG71557.1 JAG71810.1 GBYB01001780 JAG71547.1 KQ980824 KYN12524.1 KQ976419 KYM89268.1 ADTU01023417 NNAY01000927 OXU25860.1 KQ982021 KYN30587.1 ACPB03005173 LJIG01022876 KRT78277.1 GL435030 EFN74221.1 KQ982335 KYQ57547.1 GL446605 EFN87513.1 GEDC01029504 GEDC01023686 JAS07794.1 JAS13612.1 GEDC01020921 GEDC01009377 JAS16377.1 JAS27921.1 KZ288256 PBC30603.1 GEZM01078343 JAV62760.1 GDKW01001265 JAI55330.1 KQ760132 OAD61801.1 KQ414654 KOC66019.1 APGK01052521 APGK01052522 KB741213 ENN72599.1 BT128446 AEE63403.1 GECZ01015952 JAS53817.1 GBGD01000859 JAC88030.1 GECU01026144 GECU01002689 JAS81562.1 JAT05018.1 GBYB01014977 JAG84744.1 GECU01027543 GECU01014113 JAS80163.1 JAS93593.1 GGMS01016473 MBY85676.1 GECL01001806 JAP04318.1 GFXV01000071 MBW11876.1 ABLF02039071 KQ435712 KOX79357.1 KQ434923 KZC11486.1 LBMM01000464 KMQ98275.1 DS235171 EEB12905.1 GAIX01004484 JAA88076.1 AJVK01010231 GBHO01016137 GBHO01016136 GBHO01016133 GBHO01005438 GBRD01008408 JAG27467.1 JAG27468.1 JAG27471.1 JAG38166.1 JAG57413.1 GFDF01002404 JAV11680.1 JH431669 JXUM01103444 KQ564805 KXJ71726.1 GDUN01000683 JAN95236.1 GDIP01095846 JAM07869.1 GDIP01149841 GDIQ01035029 LRGB01000005 JAJ73561.1 JAN59708.1 KZS22012.1 GGFK01006964 MBW40285.1 GFDL01001615 JAV33430.1 GGFK01006899 MBW40220.1 GDIP01214080 JAJ09322.1 AXCN02002502 UFQS01000093 UFQT01000093 SSW99222.1 SSX19602.1 AAAB01008839 AAAB01008980 APCN01004541 EAA14432.3 ATLV01020113 KE525294 KFB44856.1 ADMH02002162 ETN58165.1 GDIQ01019032 JAN75705.1 GECZ01026257 JAS43512.1 AMQN01006822 KB299236 ELU08223.1 AXCM01004598 KA648128 AFP62757.1 JRES01000699 KNC29075.1 GAKP01023438 JAC35520.1 GDHF01021234 JAI31080.1 JXJN01023173 GAMC01006727 JAB99828.1 GDIQ01070204 JAN24533.1 CVRI01000001 CRK86447.1 CCAG010007366 GBXI01008555 JAD05737.1
Proteomes
UP000218220
UP000053240
UP000283053
UP000235965
UP000078542
UP000027135
+ More
UP000053097 UP000279307 UP000007266 UP000192223 UP000078492 UP000078540 UP000002358 UP000005205 UP000215335 UP000078541 UP000015103 UP000000311 UP000075809 UP000008237 UP000005203 UP000242457 UP000053825 UP000019118 UP000079169 UP000007819 UP000053105 UP000076502 UP000036403 UP000009046 UP000092462 UP000069940 UP000249989 UP000076858 UP000075881 UP000075886 UP000075920 UP000075900 UP000075884 UP000075880 UP000075885 UP000075903 UP000075902 UP000076407 UP000075882 UP000075840 UP000007062 UP000030765 UP000069272 UP000000673 UP000095301 UP000014760 UP000075883 UP000037069 UP000092443 UP000092460 UP000078200 UP000092445 UP000183832 UP000092444
UP000053097 UP000279307 UP000007266 UP000192223 UP000078492 UP000078540 UP000002358 UP000005205 UP000215335 UP000078541 UP000015103 UP000000311 UP000075809 UP000008237 UP000005203 UP000242457 UP000053825 UP000019118 UP000079169 UP000007819 UP000053105 UP000076502 UP000036403 UP000009046 UP000092462 UP000069940 UP000249989 UP000076858 UP000075881 UP000075886 UP000075920 UP000075900 UP000075884 UP000075880 UP000075885 UP000075903 UP000075902 UP000076407 UP000075882 UP000075840 UP000007062 UP000030765 UP000069272 UP000000673 UP000095301 UP000014760 UP000075883 UP000037069 UP000092443 UP000092460 UP000078200 UP000092445 UP000183832 UP000092444
Interpro
SUPFAM
SSF52799
SSF52799
Gene 3D
ProteinModelPortal
A0A2H1WKH1
A0A2A4JBE7
A0A194RDN7
A0A437B033
A0A2J7Q1C2
A0A195D504
+ More
A0A067R4P3 A0A026WLL8 A0A3L8E7Q5 A0A1B6MD37 A0A1B6L6A7 D6W9S3 A0A0C9QN08 A0A0C9R1G5 A0A0C9R1G2 A0A1W4X484 A0A195DJ47 A0A195BQW0 K7ITS3 A0A158NQM9 A0A232F5U2 A0A195ERR0 T1I1D3 A0A0T6ATR8 E1ZXD1 A0A151XB23 E2B9S6 A0A1B6CJN7 A0A1B6CSM6 A0A088ACG1 A0A2A3EFN7 A0A1Y1KRF7 A0A0P4VRX4 A0A310STE7 A0A0L7R5C2 N6SXW9 J3JYW5 A0A1B6FUB7 A0A069DVU4 A0A1B6K0N9 A0A0C9RNB0 A0A1B6HZS2 A0A1S3CXK2 A0A2S2R6L7 A0A0V0G8M7 A0A2H8TCN2 J9JNR1 A0A0M9A8G1 A0A154PHY4 A0A0J7P0V1 E0VHL9 S4P7Q9 A0A1B0D1E8 A0A0A9Y770 A0A1L8DYY1 T1IY29 A0A182H1G5 A0A0P6J551 A0A0P5VTG8 A0A0N8AGG2 A0A2M4AHL3 A0A1Q3G0W1 A0A182JVD7 A0A2M4AHG4 A0A0P4ZNA5 A0A182QW07 A0A182VV39 A0A182R9C9 A0A182NLJ6 A0A182JG63 A0A336K5U4 A0A1S4H1F2 A0A182PB34 A0A182VBA3 A0A182TZ48 A0A182XCL7 A0A182L5V7 A0A182IDZ3 Q7Q1Z5 A0A084W3R2 A0A182F216 W5J5L9 A0A0P6HGU3 A0A1I8M937 A0A1B6EZR2 R7UQ41 A0A182M6M2 T1PJL1 A0A0L0CA33 A0A034V144 A0A0K8UXI6 A0A1A9YDW5 A0A1B0BZJ2 A0A1A9UNK3 A0A1B0A5M0 W8BSE7 A0A0N8E1T3 A0A1J1HER9 A0A1B0FDL8 A0A0A1X3Q1
A0A067R4P3 A0A026WLL8 A0A3L8E7Q5 A0A1B6MD37 A0A1B6L6A7 D6W9S3 A0A0C9QN08 A0A0C9R1G5 A0A0C9R1G2 A0A1W4X484 A0A195DJ47 A0A195BQW0 K7ITS3 A0A158NQM9 A0A232F5U2 A0A195ERR0 T1I1D3 A0A0T6ATR8 E1ZXD1 A0A151XB23 E2B9S6 A0A1B6CJN7 A0A1B6CSM6 A0A088ACG1 A0A2A3EFN7 A0A1Y1KRF7 A0A0P4VRX4 A0A310STE7 A0A0L7R5C2 N6SXW9 J3JYW5 A0A1B6FUB7 A0A069DVU4 A0A1B6K0N9 A0A0C9RNB0 A0A1B6HZS2 A0A1S3CXK2 A0A2S2R6L7 A0A0V0G8M7 A0A2H8TCN2 J9JNR1 A0A0M9A8G1 A0A154PHY4 A0A0J7P0V1 E0VHL9 S4P7Q9 A0A1B0D1E8 A0A0A9Y770 A0A1L8DYY1 T1IY29 A0A182H1G5 A0A0P6J551 A0A0P5VTG8 A0A0N8AGG2 A0A2M4AHL3 A0A1Q3G0W1 A0A182JVD7 A0A2M4AHG4 A0A0P4ZNA5 A0A182QW07 A0A182VV39 A0A182R9C9 A0A182NLJ6 A0A182JG63 A0A336K5U4 A0A1S4H1F2 A0A182PB34 A0A182VBA3 A0A182TZ48 A0A182XCL7 A0A182L5V7 A0A182IDZ3 Q7Q1Z5 A0A084W3R2 A0A182F216 W5J5L9 A0A0P6HGU3 A0A1I8M937 A0A1B6EZR2 R7UQ41 A0A182M6M2 T1PJL1 A0A0L0CA33 A0A034V144 A0A0K8UXI6 A0A1A9YDW5 A0A1B0BZJ2 A0A1A9UNK3 A0A1B0A5M0 W8BSE7 A0A0N8E1T3 A0A1J1HER9 A0A1B0FDL8 A0A0A1X3Q1
PDB
1M7R
E-value=0,
Score=1799
Ontologies
PATHWAY
GO
PANTHER
Topology
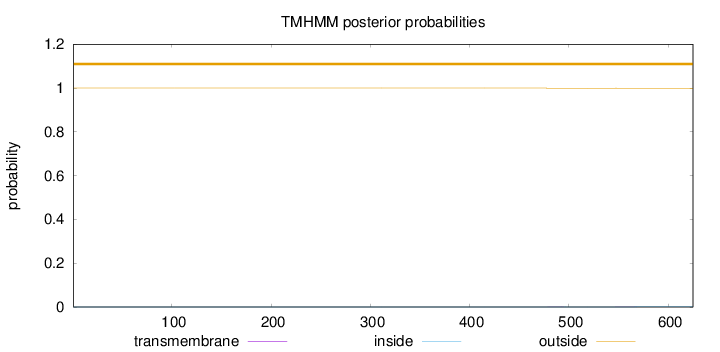
Length:
625
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01295
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00031
outside
1 - 625
Population Genetic Test Statistics
Pi
207.131327
Theta
18.289912
Tajima's D
-1.390869
CLR
1407.541808
CSRT
0.0751462426878656
Interpretation
Uncertain
