Gene
KWMTBOMO08846
Annotation
PREDICTED:_chondroitin_proteoglycan_2-like_[Plutella_xylostella]
Full name
Protein obstructor-E
Location in the cell
Extracellular Reliability : 4.349
Sequence
CDS
ATGCTCTCCTACATCTACCTCGCCTTGCTAGTGGCTGCAAGCCATGTCCAATCAACTCCCGCTGCTGCTACTGCCCAGAACGTAATCTGCAAAGCCCGCAATGGTTATTACAAAACAGACGCGAATTGTGATACATACATTGAATGCCGGGACTATCAAGCTACTAACATGATATGCCCGGATGGTCTGCACTTTAATCCTAGCGTGGAATGGCCGGCATATCCTTGCGGTTATCCAGTTGAAGTGACATGTGTAGGAAGAGGGAGCATCCAGCCTGCCCAAACAACACCGGATTGCCCGCACCAATATGGCATCTTCAAACATCCTAACGCTTCGCCCACCAATTGTGGCCAGTACCGAACATGCGTAGGTGGTCGTGCTTTCGACATGGTATGCCCGCCGGGACTAGCATTCAATCCAGACTTCAGCCGCTGTGACTGGGCCGACCTTGTACCTTCCTGTGATGCTGAAAAATTCTTGGGTTTCACTTGCCCCCCTGCTCCTTTGGATGCTATCGGCAATCCTCTGAATAATGTGATAAACTACAAGTACGAAGGCAACTGCTACTACTTCTTCTCCTGTGAGCAAAATCGGGCTAGATTGCTGTCCTGCGACATCGGTCTTGCATTCGACCCCACGACTGGACGGTGCGTCGACGCAGACAGAGTCCAGTGCAATGCAACGCAAAACACAAATGACAAGAATCATATATGA
Protein
MLSYIYLALLVAASHVQSTPAAATAQNVICKARNGYYKTDANCDTYIECRDYQATNMICPDGLHFNPSVEWPAYPCGYPVEVTCVGRGSIQPAQTTPDCPHQYGIFKHPNASPTNCGQYRTCVGGRAFDMVCPPGLAFNPDFSRCDWADLVPSCDAEKFLGFTCPPAPLDAIGNPLNNVINYKYEGNCYYFFSCEQNRARLLSCDIGLAFDPTTGRCVDADRVQCNATQNTNDKNHI
Summary
Description
Chitin-binding protein that is important for the longitudinal contraction and lateral expansion of the larval cuticle during its conversion into the oval-shaped puparium case. Essential for survival to the second instar larval stage. Confers the orientated contractility and expandability of the larval cuticle by regulating the arrangement of chitin and the formation of supracellular ridges on the cuticle of third instar larvae. Essential for determining pupal body shape; required for the orientated shape change of the cuticle during metamorphosis which involves changes in the morphology of the supracellular ridges.
Isoform A: Mainly involved in regulating pupal shape.
Isoform B: Mainly involved in larvae survival, possibly by maintaining the normal morphology of the larval hindgut during development.
Isoform A: Mainly involved in regulating pupal shape.
Isoform B: Mainly involved in larvae survival, possibly by maintaining the normal morphology of the larval hindgut during development.
Keywords
Alternative splicing
Chitin-binding
Complete proteome
Cuticle
Disulfide bond
Extracellular matrix
Glycoprotein
Reference proteome
Repeat
Secreted
Signal
Feature
chain Protein obstructor-E
splice variant In isoform A.
splice variant In isoform A.
Uniprot
A0A2W1BF23
A0A2A4JBU2
A0A3S2N7I4
A0A212F4E7
A0A212F4E4
A0A194PQG3
+ More
A0A194PWP1 A0A194PS25 A0A194RE65 A0A194PQ33 A0A194RDP2 A0A212F4D0 A0A194QP38 I4DML9 A0A194PQ28 I4DNE9 A0A0L7KUM0 A0A0F6PN10 A0A1B6D857 K7ILZ1 A0A3S2LC51 A0A3S2LTG2 D1MAI7 A0A1Y1N6X3 A0A026W6E9 A0A0F6PMG7 E9JDK5 A0A151X4R9 A0A195BCC5 A0A2J7RMS5 A0A195FXS8 A0A195DE51 F4WJU2 A0A158NS31 A0A151ILF7 A0A067RK54 A0A0A1WW87 A0A034W0W1 E2AYK3 A0A2S1ZS46 A0A2H1WIQ5 A0A2A4JAB4 A0A1B0D1P0 A0A023F905 A0A0K8V2U4 A0A212F4E1 A0A0C9QB70 A0A2Y9D4S8 A0A2W1B9A5 A0A224XY50 A0A1I8MZF1 A0A2M4BZ99 R4FMJ4 W8C6S2 A0A182IT75 J3JX93 A0A2S2R3N9 A0A0L7R0S2 U4UB08 N6TSA9 A0A1I8N1E5 A0A1D2MQI6 A0A2H8TE74 A0A154PAJ4 E0VLN7 A0A2S2PMI9 A0A146M7V2 A0A0A9VVQ8 A0A0L0BWY5 W5J7F5 A0A182VUQ9 A0A336MRH4 A0A2A3ED73 A0A182F1V8 A0A0K8SP49 A0A226F411 A0A1W4W557 B4M9J0 J9JK90 T1DNE2 A0A310S848 A0A088AMM7 B3MMM2 A0A2R7VZS5 A0A182RG78 B4JC70 A0A0K8SQI3 A0A0J9QWD0 B4I1K3 A0A0R1DTS6 Q9VMM6-2 A0A182SDT0 A0A182KBC6 A0A182YFW5 B4KGZ8 A0A0Q5WIN5 A0A182QV04 E2C2F8 A0A1J1HSH1
A0A194PWP1 A0A194PS25 A0A194RE65 A0A194PQ33 A0A194RDP2 A0A212F4D0 A0A194QP38 I4DML9 A0A194PQ28 I4DNE9 A0A0L7KUM0 A0A0F6PN10 A0A1B6D857 K7ILZ1 A0A3S2LC51 A0A3S2LTG2 D1MAI7 A0A1Y1N6X3 A0A026W6E9 A0A0F6PMG7 E9JDK5 A0A151X4R9 A0A195BCC5 A0A2J7RMS5 A0A195FXS8 A0A195DE51 F4WJU2 A0A158NS31 A0A151ILF7 A0A067RK54 A0A0A1WW87 A0A034W0W1 E2AYK3 A0A2S1ZS46 A0A2H1WIQ5 A0A2A4JAB4 A0A1B0D1P0 A0A023F905 A0A0K8V2U4 A0A212F4E1 A0A0C9QB70 A0A2Y9D4S8 A0A2W1B9A5 A0A224XY50 A0A1I8MZF1 A0A2M4BZ99 R4FMJ4 W8C6S2 A0A182IT75 J3JX93 A0A2S2R3N9 A0A0L7R0S2 U4UB08 N6TSA9 A0A1I8N1E5 A0A1D2MQI6 A0A2H8TE74 A0A154PAJ4 E0VLN7 A0A2S2PMI9 A0A146M7V2 A0A0A9VVQ8 A0A0L0BWY5 W5J7F5 A0A182VUQ9 A0A336MRH4 A0A2A3ED73 A0A182F1V8 A0A0K8SP49 A0A226F411 A0A1W4W557 B4M9J0 J9JK90 T1DNE2 A0A310S848 A0A088AMM7 B3MMM2 A0A2R7VZS5 A0A182RG78 B4JC70 A0A0K8SQI3 A0A0J9QWD0 B4I1K3 A0A0R1DTS6 Q9VMM6-2 A0A182SDT0 A0A182KBC6 A0A182YFW5 B4KGZ8 A0A0Q5WIN5 A0A182QV04 E2C2F8 A0A1J1HSH1
Pubmed
28756777
22118469
26354079
22651552
26227816
20075255
+ More
18362917 20144715 19820115 28004739 24508170 21282665 21719571 21347285 24845553 25830018 25348373 20798317 29712872 25474469 25315136 24495485 22516182 23537049 27289101 20566863 26823975 25401762 26108605 20920257 23761445 17994087 22936249 17550304 10731132 12537572 12537569 28076349 25244985
18362917 20144715 19820115 28004739 24508170 21282665 21719571 21347285 24845553 25830018 25348373 20798317 29712872 25474469 25315136 24495485 22516182 23537049 27289101 20566863 26823975 25401762 26108605 20920257 23761445 17994087 22936249 17550304 10731132 12537572 12537569 28076349 25244985
EMBL
KZ150252
PZC71807.1
NWSH01002160
PCG69024.1
RSAL01000249
RVE43503.1
+ More
AGBW02010386 OWR48593.1 OWR48592.1 KQ459596 KPI95552.1 KPI95555.1 KPI95554.1 KQ460367 KPJ15575.1 KPI95556.1 KPJ15574.1 OWR48591.1 KQ461195 KPJ06740.1 AK402537 BAM19159.1 KPI95551.1 AK402874 BAM19439.1 JTDY01005687 KOB66751.1 KM099164 AJZ68825.1 GEDC01015415 JAS21883.1 RVE43501.1 RVE43504.1 GU128095 KQ971379 ACY95478.1 EEZ97505.1 GEZM01014692 JAV92036.1 KK107374 EZA51588.1 KM099165 AJZ68826.1 GL771873 EFZ09095.1 KQ982544 KYQ55351.1 KQ976528 KYM81872.1 NEVH01002552 PNF42122.1 KQ981208 KYN44649.1 KQ980989 KYN10704.1 GL888186 EGI65584.1 ADTU01024435 KQ977142 KYN05321.1 KK852463 KDR23408.1 GBXI01011522 JAD02770.1 GAKP01011200 GAKP01011197 JAC47755.1 GL443918 EFN61491.1 MF942766 AWK28289.1 ODYU01008934 SOQ52949.1 PCG69025.1 AJVK01010349 GBBI01001268 JAC17444.1 GDHF01019097 JAI33217.1 OWR48594.1 GBYB01000481 GBYB01000484 JAG70248.1 JAG70251.1 PZC71808.1 GFTR01003417 JAW13009.1 GGFJ01009201 MBW58342.1 ACPB03017897 GAHY01001694 JAA75816.1 GAMC01004089 JAC02467.1 BT127862 AEE62824.1 GGMS01015382 MBY84585.1 KQ414670 KOC64447.1 KB632275 ERL91099.1 APGK01053934 KB741239 ENN72125.1 LJIJ01000684 ODM95337.1 GFXV01000584 MBW12389.1 KQ434857 KZC08861.1 DS235277 EEB14293.1 GGMR01018043 MBY30662.1 GDHC01003793 JAQ14836.1 GBHO01044323 JAF99280.1 JRES01001211 KNC24558.1 ADMH02002118 ETN58800.1 UFQS01000186 UFQS01002158 UFQT01000186 UFQT01002158 SSX00899.1 SSX32816.1 KZ288280 PBC29660.1 GBRD01010738 JAG55086.1 LNIX01000001 OXA64509.1 CH940654 EDW57866.1 ABLF02021901 ABLF02021903 GAMD01002960 JAA98630.1 KQ764887 OAD54244.1 CH902620 EDV31913.1 KK854198 PTY12972.1 CH916368 EDW03083.1 GBRD01010739 JAG55085.1 CM002910 KMY88366.1 CH480820 EDW54410.1 CM000157 KRJ98086.1 AE014134 AY069031 BT099959 BT001600 AAL39176.1 AAN71355.1 ACX47660.1 CH933807 EDW12209.1 CH954177 KQS70190.1 AXCN02000847 GL452124 EFN77908.1 CVRI01000020 CRK90931.1
AGBW02010386 OWR48593.1 OWR48592.1 KQ459596 KPI95552.1 KPI95555.1 KPI95554.1 KQ460367 KPJ15575.1 KPI95556.1 KPJ15574.1 OWR48591.1 KQ461195 KPJ06740.1 AK402537 BAM19159.1 KPI95551.1 AK402874 BAM19439.1 JTDY01005687 KOB66751.1 KM099164 AJZ68825.1 GEDC01015415 JAS21883.1 RVE43501.1 RVE43504.1 GU128095 KQ971379 ACY95478.1 EEZ97505.1 GEZM01014692 JAV92036.1 KK107374 EZA51588.1 KM099165 AJZ68826.1 GL771873 EFZ09095.1 KQ982544 KYQ55351.1 KQ976528 KYM81872.1 NEVH01002552 PNF42122.1 KQ981208 KYN44649.1 KQ980989 KYN10704.1 GL888186 EGI65584.1 ADTU01024435 KQ977142 KYN05321.1 KK852463 KDR23408.1 GBXI01011522 JAD02770.1 GAKP01011200 GAKP01011197 JAC47755.1 GL443918 EFN61491.1 MF942766 AWK28289.1 ODYU01008934 SOQ52949.1 PCG69025.1 AJVK01010349 GBBI01001268 JAC17444.1 GDHF01019097 JAI33217.1 OWR48594.1 GBYB01000481 GBYB01000484 JAG70248.1 JAG70251.1 PZC71808.1 GFTR01003417 JAW13009.1 GGFJ01009201 MBW58342.1 ACPB03017897 GAHY01001694 JAA75816.1 GAMC01004089 JAC02467.1 BT127862 AEE62824.1 GGMS01015382 MBY84585.1 KQ414670 KOC64447.1 KB632275 ERL91099.1 APGK01053934 KB741239 ENN72125.1 LJIJ01000684 ODM95337.1 GFXV01000584 MBW12389.1 KQ434857 KZC08861.1 DS235277 EEB14293.1 GGMR01018043 MBY30662.1 GDHC01003793 JAQ14836.1 GBHO01044323 JAF99280.1 JRES01001211 KNC24558.1 ADMH02002118 ETN58800.1 UFQS01000186 UFQS01002158 UFQT01000186 UFQT01002158 SSX00899.1 SSX32816.1 KZ288280 PBC29660.1 GBRD01010738 JAG55086.1 LNIX01000001 OXA64509.1 CH940654 EDW57866.1 ABLF02021901 ABLF02021903 GAMD01002960 JAA98630.1 KQ764887 OAD54244.1 CH902620 EDV31913.1 KK854198 PTY12972.1 CH916368 EDW03083.1 GBRD01010739 JAG55085.1 CM002910 KMY88366.1 CH480820 EDW54410.1 CM000157 KRJ98086.1 AE014134 AY069031 BT099959 BT001600 AAL39176.1 AAN71355.1 ACX47660.1 CH933807 EDW12209.1 CH954177 KQS70190.1 AXCN02000847 GL452124 EFN77908.1 CVRI01000020 CRK90931.1
Proteomes
UP000218220
UP000283053
UP000007151
UP000053268
UP000053240
UP000037510
+ More
UP000002358 UP000007266 UP000053097 UP000075809 UP000078540 UP000235965 UP000078541 UP000078492 UP000007755 UP000005205 UP000078542 UP000027135 UP000000311 UP000092462 UP000095300 UP000095301 UP000015103 UP000075880 UP000053825 UP000030742 UP000019118 UP000094527 UP000076502 UP000009046 UP000037069 UP000000673 UP000075920 UP000242457 UP000069272 UP000198287 UP000192223 UP000008792 UP000007819 UP000005203 UP000007801 UP000075900 UP000001070 UP000001292 UP000002282 UP000000803 UP000075901 UP000075881 UP000076408 UP000009192 UP000008711 UP000075886 UP000008237 UP000183832
UP000002358 UP000007266 UP000053097 UP000075809 UP000078540 UP000235965 UP000078541 UP000078492 UP000007755 UP000005205 UP000078542 UP000027135 UP000000311 UP000092462 UP000095300 UP000095301 UP000015103 UP000075880 UP000053825 UP000030742 UP000019118 UP000094527 UP000076502 UP000009046 UP000037069 UP000000673 UP000075920 UP000242457 UP000069272 UP000198287 UP000192223 UP000008792 UP000007819 UP000005203 UP000007801 UP000075900 UP000001070 UP000001292 UP000002282 UP000000803 UP000075901 UP000075881 UP000076408 UP000009192 UP000008711 UP000075886 UP000008237 UP000183832
PRIDE
Pfam
PF01607 CBM_14
SUPFAM
SSF57625
SSF57625
ProteinModelPortal
A0A2W1BF23
A0A2A4JBU2
A0A3S2N7I4
A0A212F4E7
A0A212F4E4
A0A194PQG3
+ More
A0A194PWP1 A0A194PS25 A0A194RE65 A0A194PQ33 A0A194RDP2 A0A212F4D0 A0A194QP38 I4DML9 A0A194PQ28 I4DNE9 A0A0L7KUM0 A0A0F6PN10 A0A1B6D857 K7ILZ1 A0A3S2LC51 A0A3S2LTG2 D1MAI7 A0A1Y1N6X3 A0A026W6E9 A0A0F6PMG7 E9JDK5 A0A151X4R9 A0A195BCC5 A0A2J7RMS5 A0A195FXS8 A0A195DE51 F4WJU2 A0A158NS31 A0A151ILF7 A0A067RK54 A0A0A1WW87 A0A034W0W1 E2AYK3 A0A2S1ZS46 A0A2H1WIQ5 A0A2A4JAB4 A0A1B0D1P0 A0A023F905 A0A0K8V2U4 A0A212F4E1 A0A0C9QB70 A0A2Y9D4S8 A0A2W1B9A5 A0A224XY50 A0A1I8MZF1 A0A2M4BZ99 R4FMJ4 W8C6S2 A0A182IT75 J3JX93 A0A2S2R3N9 A0A0L7R0S2 U4UB08 N6TSA9 A0A1I8N1E5 A0A1D2MQI6 A0A2H8TE74 A0A154PAJ4 E0VLN7 A0A2S2PMI9 A0A146M7V2 A0A0A9VVQ8 A0A0L0BWY5 W5J7F5 A0A182VUQ9 A0A336MRH4 A0A2A3ED73 A0A182F1V8 A0A0K8SP49 A0A226F411 A0A1W4W557 B4M9J0 J9JK90 T1DNE2 A0A310S848 A0A088AMM7 B3MMM2 A0A2R7VZS5 A0A182RG78 B4JC70 A0A0K8SQI3 A0A0J9QWD0 B4I1K3 A0A0R1DTS6 Q9VMM6-2 A0A182SDT0 A0A182KBC6 A0A182YFW5 B4KGZ8 A0A0Q5WIN5 A0A182QV04 E2C2F8 A0A1J1HSH1
A0A194PWP1 A0A194PS25 A0A194RE65 A0A194PQ33 A0A194RDP2 A0A212F4D0 A0A194QP38 I4DML9 A0A194PQ28 I4DNE9 A0A0L7KUM0 A0A0F6PN10 A0A1B6D857 K7ILZ1 A0A3S2LC51 A0A3S2LTG2 D1MAI7 A0A1Y1N6X3 A0A026W6E9 A0A0F6PMG7 E9JDK5 A0A151X4R9 A0A195BCC5 A0A2J7RMS5 A0A195FXS8 A0A195DE51 F4WJU2 A0A158NS31 A0A151ILF7 A0A067RK54 A0A0A1WW87 A0A034W0W1 E2AYK3 A0A2S1ZS46 A0A2H1WIQ5 A0A2A4JAB4 A0A1B0D1P0 A0A023F905 A0A0K8V2U4 A0A212F4E1 A0A0C9QB70 A0A2Y9D4S8 A0A2W1B9A5 A0A224XY50 A0A1I8MZF1 A0A2M4BZ99 R4FMJ4 W8C6S2 A0A182IT75 J3JX93 A0A2S2R3N9 A0A0L7R0S2 U4UB08 N6TSA9 A0A1I8N1E5 A0A1D2MQI6 A0A2H8TE74 A0A154PAJ4 E0VLN7 A0A2S2PMI9 A0A146M7V2 A0A0A9VVQ8 A0A0L0BWY5 W5J7F5 A0A182VUQ9 A0A336MRH4 A0A2A3ED73 A0A182F1V8 A0A0K8SP49 A0A226F411 A0A1W4W557 B4M9J0 J9JK90 T1DNE2 A0A310S848 A0A088AMM7 B3MMM2 A0A2R7VZS5 A0A182RG78 B4JC70 A0A0K8SQI3 A0A0J9QWD0 B4I1K3 A0A0R1DTS6 Q9VMM6-2 A0A182SDT0 A0A182KBC6 A0A182YFW5 B4KGZ8 A0A0Q5WIN5 A0A182QV04 E2C2F8 A0A1J1HSH1
PDB
6G9E
E-value=8.1171e-05,
Score=107
Ontologies
GO
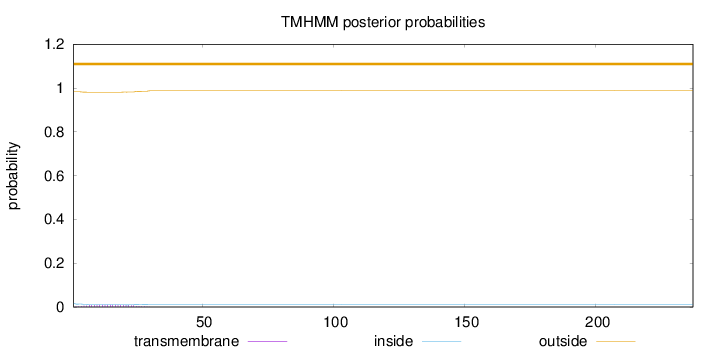
Topology
Subcellular location
SignalP
Position: 1 - 18,
Likelihood: 0.997355
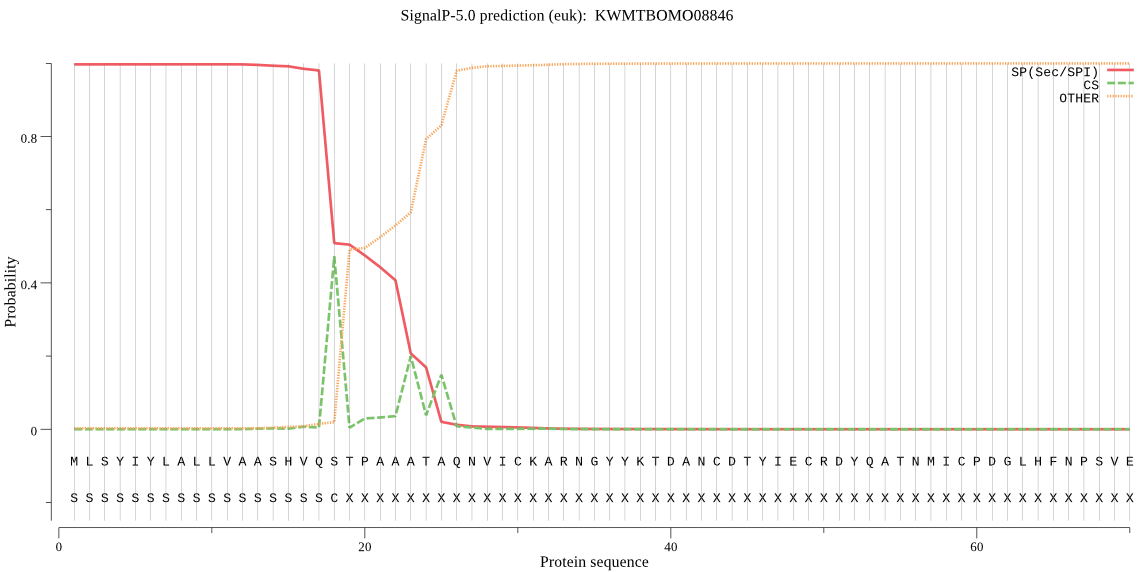
Length:
237
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.20277
Exp number, first 60 AAs:
0.20239
Total prob of N-in:
0.01648
outside
1 - 237
Population Genetic Test Statistics
Pi
169.882764
Theta
211.736028
Tajima's D
-0.773702
CLR
0
CSRT
0.184690765461727
Interpretation
Uncertain
