Gene
KWMTBOMO08841
Pre Gene Modal
BGIBMGA007852
Annotation
PREDICTED:_carbohydrate_sulfotransferase_3-like_isoform_X2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.596 Mitochondrial Reliability : 1.543 Nuclear Reliability : 1.268
Sequence
CDS
ATGGCTGCAGGCGTAAACGTTGTTAAGCTCGCTGGTAATTTATTTCGCAGCAAAATATCGTTGTTCCAACAATGGACAGAAGTAAATATGCCACTCAAAAGCAAAGAAAAAATAAAGGATTTGTTGCAAGAGCAAATCGAAAACGCAAAAAGATCAAAACTTGCTGAGAAAATGAAGACCGCGAAAGGTTTTTACACCCTGGAAATGGCACCACCATCAAGCTCAGAGGAAATGGCCAAACTTAAAGAAGATTTGAAATTATTTCAAGAATTCATGAAATCTGGCTGCTACAAACACCTGACAGTCAAACAAGCGTGGCTTCTCTTCTTGATTAGTACGGAAATCGGCCTATGGTTCTTCTTGGGCGAAACGATTGGGAAATTTCACATCGTTGGCTATCAACCGGTACCTACGAATTCTTACGATGGAACACCGAATATAGAAAAAATATTAGAAAAAACACGAGCTAAAATTCAATTTGAATTGTGGAACTATAACTTTACTTATTCTGGAGTTAAAAAATTAGATGACTTAATTATAGAAACCGGTGGTCGTCCATTGAGAAGTATCATAATATCTTCTTGGAGATCTGGTTCTACATTCCTGGATTACGGACGAGGACATAACCATCAGTTCAGTCATAACACCAGATTATGGGACCATTGTAAATATAAGAAGGAACTTTGTTTTGATTCTGATTTTATGTCAAAATTTTGCAAATTATTTCCATTTCAAAGTATGAAAGTTGTTCGCCTTAGATTGCGTCTTGTTCAGGAATTGTTGGAAGATAAAGAGCTTAATGCAAAAGTTGTGTTTTTAATCCGAGATCCTCGTGGTGTCCTACAGTCAAGATTGCATCGTGATTTCTGTCACCCTTCACCAGATTGTTCGAATCCTGAGCTCCTCTGTGCAGACATGATAAGTGATTATGTGGCTGCGAGCCGATTGCTCAAGCTTTACCCTGATAAACTTATGGTGCTACGCTATGAAGACTTGGCCCTGAATCTAAATAGTACCACGCACAAATTATTAAAGTTCCTAAGACTGAGTCACATGCAAGCTGTTGATGAGTATCTAAACTCTCACACCAACACAGAAGTGAGCGGAGTAAGCTCCACTTTTAGAGTATCCCGTGAAGTGCCTTTTCGGTGGAAGAATATATTGGATTTTGAATATGCTGATCAAATACAGATGGCGTGTAAAGAAGCCATGAATTTATGGGGTTATCGTATGGCTCACAACGCCACCCATATGACAAGCAAAGAATTTGATCCTTTAGAAACATATACGATAACTCAGTGA
Protein
MAAGVNVVKLAGNLFRSKISLFQQWTEVNMPLKSKEKIKDLLQEQIENAKRSKLAEKMKTAKGFYTLEMAPPSSSEEMAKLKEDLKLFQEFMKSGCYKHLTVKQAWLLFLISTEIGLWFFLGETIGKFHIVGYQPVPTNSYDGTPNIEKILEKTRAKIQFELWNYNFTYSGVKKLDDLIIETGGRPLRSIIISSWRSGSTFLDYGRGHNHQFSHNTRLWDHCKYKKELCFDSDFMSKFCKLFPFQSMKVVRLRLRLVQELLEDKELNAKVVFLIRDPRGVLQSRLHRDFCHPSPDCSNPELLCADMISDYVAASRLLKLYPDKLMVLRYEDLALNLNSTTHKLLKFLRLSHMQAVDEYLNSHTNTEVSGVSSTFRVSREVPFRWKNILDFEYADQIQMACKEAMNLWGYRMAHNATHMTSKEFDPLETYTITQ
Summary
Uniprot
H9JEA5
A0A194PQF8
A0A437AZA0
A0A0L7KTC0
A0A2A4JKF2
A0A2W1BJH7
+ More
A0A1E1W9V6 H9J4S9 A0A212F4E5 A0A154P009 A0A0M8ZY24 A0A0L7RAF8 A0A088ASZ0 A0A195CRL0 A0A151IT61 E2B3G5 A0A151XA79 A0A310SHV7 A0A195FWP7 A0A195AUY0 A0A158NNA0 F4WLI7 E2A541 A0A3L8DGB5 A0A023FAA7 A0A224XQX1 A0A026WSS9 A0A069DTA0 A0A232EKU0 A0A1S4G7A8 A0A0J7KE25 A0A0N7Z9B7 A0A1Q3FTK8 A0A0M4EH57 E9IUE8 B4KKW7 A0A2W1BH33 A0A1W4WH07 K7JC85 A0A2M4CPV5 A0A2M4CPZ3 B4JQ84 A0A2R5L1M4 A0A0A1WJQ0 B4M9L1 A0A3Q0J971 B4N7N5 A0A2M4CPS9 A0A1B0Y0C3 W8BD39 A0A0P4W187 A0A0K8WKU8 A0A034VZ02 U5EG46 D6X452 A0A2M4BNS8 A0A2M4BSG5 A0A2J7RMR0 A0A1B6DXR3 A0A1B6J9K2 T1IAB8 B3N5S9 Q9VMC3 A0A1W4V270 B4P0C5 B4Q4X9 A0A3B0KBB5 A0A067RHX8 A0A0L0C491 A0A2M4BSU4 B3MP66 Q29PD2 A0A0T6AUP2 Q7Q1F9 A0A423SVV8 T1PJH1 A0A1S4H2X8 A0A182J7N5 A0A2C9GPU3 A0A084WG47 R4FKL8 A0A182V1S9 A0A1I8MIU1 A0A1D2MXQ9 E0VHS3 A0A1B6MIT2 A0A1I8Q9U4 W5J5F5 A0A182KTW2 A0A2R7WCE1 A0A1B6CQM9 A0A226EME3 A0A182Q212
A0A1E1W9V6 H9J4S9 A0A212F4E5 A0A154P009 A0A0M8ZY24 A0A0L7RAF8 A0A088ASZ0 A0A195CRL0 A0A151IT61 E2B3G5 A0A151XA79 A0A310SHV7 A0A195FWP7 A0A195AUY0 A0A158NNA0 F4WLI7 E2A541 A0A3L8DGB5 A0A023FAA7 A0A224XQX1 A0A026WSS9 A0A069DTA0 A0A232EKU0 A0A1S4G7A8 A0A0J7KE25 A0A0N7Z9B7 A0A1Q3FTK8 A0A0M4EH57 E9IUE8 B4KKW7 A0A2W1BH33 A0A1W4WH07 K7JC85 A0A2M4CPV5 A0A2M4CPZ3 B4JQ84 A0A2R5L1M4 A0A0A1WJQ0 B4M9L1 A0A3Q0J971 B4N7N5 A0A2M4CPS9 A0A1B0Y0C3 W8BD39 A0A0P4W187 A0A0K8WKU8 A0A034VZ02 U5EG46 D6X452 A0A2M4BNS8 A0A2M4BSG5 A0A2J7RMR0 A0A1B6DXR3 A0A1B6J9K2 T1IAB8 B3N5S9 Q9VMC3 A0A1W4V270 B4P0C5 B4Q4X9 A0A3B0KBB5 A0A067RHX8 A0A0L0C491 A0A2M4BSU4 B3MP66 Q29PD2 A0A0T6AUP2 Q7Q1F9 A0A423SVV8 T1PJH1 A0A1S4H2X8 A0A182J7N5 A0A2C9GPU3 A0A084WG47 R4FKL8 A0A182V1S9 A0A1I8MIU1 A0A1D2MXQ9 E0VHS3 A0A1B6MIT2 A0A1I8Q9U4 W5J5F5 A0A182KTW2 A0A2R7WCE1 A0A1B6CQM9 A0A226EME3 A0A182Q212
Pubmed
19121390
26354079
26227816
28756777
22118469
20798317
+ More
21347285 21719571 30249741 25474469 24508170 26334808 28648823 27129103 21282665 17994087 20075255 25830018 24495485 25348373 18362917 19820115 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 22936249 24845553 26108605 15632085 12364791 24438588 25315136 27289101 20566863 20920257 23761445 20966253
21347285 21719571 30249741 25474469 24508170 26334808 28648823 27129103 21282665 17994087 20075255 25830018 24495485 25348373 18362917 19820115 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 22936249 24845553 26108605 15632085 12364791 24438588 25315136 27289101 20566863 20920257 23761445 20966253
EMBL
BABH01002828
KQ459596
KPI95547.1
RSAL01000249
RVE43508.1
JTDY01006122
+ More
KOB66269.1 NWSH01001165 PCG72299.1 KZ150252 PZC71813.1 GDQN01007329 JAT83725.1 BABH01035433 AGBW02010386 OWR48599.1 KQ434781 KZC04460.1 KQ435798 KOX73445.1 KQ414618 KOC67872.1 KQ977372 KYN03270.1 KQ981037 KYN10153.1 GL445346 EFN89717.1 KQ982351 KYQ57277.1 KQ768301 OAD53180.1 KQ981208 KYN44742.1 KQ976738 KYM75862.1 ADTU01021189 GL888208 EGI64923.1 GL436803 EFN71447.1 QOIP01000008 RLU19480.1 GBBI01000854 JAC17858.1 GFTR01005883 JAW10543.1 KK107109 EZA59087.1 GBGD01002022 JAC86867.1 NNAY01003711 OXU18970.1 LBMM01009033 KMQ88446.1 GDKW01000995 JAI55600.1 GFDL01004106 JAV30939.1 CP012523 ALC40205.1 GL765974 EFZ15802.1 CH933807 EDW11697.1 KZ150220 PZC72110.1 AAZX01008730 AAZX01013956 GGFL01003155 MBW67333.1 GGFL01003157 MBW67335.1 CH916372 EDV99064.1 IACF01008011 LAB73594.1 GBXI01015028 JAC99263.1 CH940654 EDW57887.1 CH964214 EDW80374.1 GGFL01003156 MBW67334.1 KT000411 ANI86009.1 GAMC01015499 JAB91056.1 GDRN01092452 JAI60106.1 GDHF01000558 JAI51756.1 GAKP01011303 JAC47649.1 GANO01003598 JAB56273.1 KQ971379 EEZ97506.1 GGFJ01005564 MBW54705.1 GGFJ01006886 MBW56027.1 NEVH01002552 PNF42123.1 GEDC01006830 JAS30468.1 GECU01030951 GECU01011815 JAS76755.1 JAS95891.1 ACPB03008218 CH954177 EDV59088.1 AE014134 AY058647 AAF52398.2 AAL13876.1 CM000157 EDW87883.1 CM000361 CM002910 EDX03971.1 KMY88539.1 OUUW01000006 SPP82351.1 KK852463 KDR23407.1 JRES01000933 KNC27105.1 GGFJ01006989 MBW56130.1 CH902620 EDV32185.2 CH379058 EAL34361.2 LJIG01022773 KRT78793.1 AAAB01008980 EAA13965.4 QCYY01002671 ROT68406.1 KA648844 AFP63473.1 APCN01001736 APCN01001737 ATLV01023441 ATLV01023442 KE525343 KFB49191.1 GAHY01002424 JAA75086.1 LJIJ01000428 ODM97584.1 DS235171 EEB12846.1 GEBQ01004182 JAT35795.1 ADMH02002127 ETN58633.1 KK854611 PTY17342.1 GEDC01021614 JAS15684.1 LNIX01000003 OXA58368.1 AXCN02000692
KOB66269.1 NWSH01001165 PCG72299.1 KZ150252 PZC71813.1 GDQN01007329 JAT83725.1 BABH01035433 AGBW02010386 OWR48599.1 KQ434781 KZC04460.1 KQ435798 KOX73445.1 KQ414618 KOC67872.1 KQ977372 KYN03270.1 KQ981037 KYN10153.1 GL445346 EFN89717.1 KQ982351 KYQ57277.1 KQ768301 OAD53180.1 KQ981208 KYN44742.1 KQ976738 KYM75862.1 ADTU01021189 GL888208 EGI64923.1 GL436803 EFN71447.1 QOIP01000008 RLU19480.1 GBBI01000854 JAC17858.1 GFTR01005883 JAW10543.1 KK107109 EZA59087.1 GBGD01002022 JAC86867.1 NNAY01003711 OXU18970.1 LBMM01009033 KMQ88446.1 GDKW01000995 JAI55600.1 GFDL01004106 JAV30939.1 CP012523 ALC40205.1 GL765974 EFZ15802.1 CH933807 EDW11697.1 KZ150220 PZC72110.1 AAZX01008730 AAZX01013956 GGFL01003155 MBW67333.1 GGFL01003157 MBW67335.1 CH916372 EDV99064.1 IACF01008011 LAB73594.1 GBXI01015028 JAC99263.1 CH940654 EDW57887.1 CH964214 EDW80374.1 GGFL01003156 MBW67334.1 KT000411 ANI86009.1 GAMC01015499 JAB91056.1 GDRN01092452 JAI60106.1 GDHF01000558 JAI51756.1 GAKP01011303 JAC47649.1 GANO01003598 JAB56273.1 KQ971379 EEZ97506.1 GGFJ01005564 MBW54705.1 GGFJ01006886 MBW56027.1 NEVH01002552 PNF42123.1 GEDC01006830 JAS30468.1 GECU01030951 GECU01011815 JAS76755.1 JAS95891.1 ACPB03008218 CH954177 EDV59088.1 AE014134 AY058647 AAF52398.2 AAL13876.1 CM000157 EDW87883.1 CM000361 CM002910 EDX03971.1 KMY88539.1 OUUW01000006 SPP82351.1 KK852463 KDR23407.1 JRES01000933 KNC27105.1 GGFJ01006989 MBW56130.1 CH902620 EDV32185.2 CH379058 EAL34361.2 LJIG01022773 KRT78793.1 AAAB01008980 EAA13965.4 QCYY01002671 ROT68406.1 KA648844 AFP63473.1 APCN01001736 APCN01001737 ATLV01023441 ATLV01023442 KE525343 KFB49191.1 GAHY01002424 JAA75086.1 LJIJ01000428 ODM97584.1 DS235171 EEB12846.1 GEBQ01004182 JAT35795.1 ADMH02002127 ETN58633.1 KK854611 PTY17342.1 GEDC01021614 JAS15684.1 LNIX01000003 OXA58368.1 AXCN02000692
Proteomes
UP000005204
UP000053268
UP000283053
UP000037510
UP000218220
UP000007151
+ More
UP000076502 UP000053105 UP000053825 UP000005203 UP000078542 UP000078492 UP000008237 UP000075809 UP000078541 UP000078540 UP000005205 UP000007755 UP000000311 UP000279307 UP000053097 UP000215335 UP000036403 UP000092553 UP000009192 UP000192223 UP000002358 UP000001070 UP000008792 UP000079169 UP000007798 UP000007266 UP000235965 UP000015103 UP000008711 UP000000803 UP000192221 UP000002282 UP000000304 UP000268350 UP000027135 UP000037069 UP000007801 UP000001819 UP000007062 UP000283509 UP000075880 UP000075840 UP000030765 UP000075903 UP000095301 UP000094527 UP000009046 UP000095300 UP000000673 UP000075882 UP000198287 UP000075886
UP000076502 UP000053105 UP000053825 UP000005203 UP000078542 UP000078492 UP000008237 UP000075809 UP000078541 UP000078540 UP000005205 UP000007755 UP000000311 UP000279307 UP000053097 UP000215335 UP000036403 UP000092553 UP000009192 UP000192223 UP000002358 UP000001070 UP000008792 UP000079169 UP000007798 UP000007266 UP000235965 UP000015103 UP000008711 UP000000803 UP000192221 UP000002282 UP000000304 UP000268350 UP000027135 UP000037069 UP000007801 UP000001819 UP000007062 UP000283509 UP000075880 UP000075840 UP000030765 UP000075903 UP000095301 UP000094527 UP000009046 UP000095300 UP000000673 UP000075882 UP000198287 UP000075886
Pfam
PF00685 Sulfotransfer_1
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
H9JEA5
A0A194PQF8
A0A437AZA0
A0A0L7KTC0
A0A2A4JKF2
A0A2W1BJH7
+ More
A0A1E1W9V6 H9J4S9 A0A212F4E5 A0A154P009 A0A0M8ZY24 A0A0L7RAF8 A0A088ASZ0 A0A195CRL0 A0A151IT61 E2B3G5 A0A151XA79 A0A310SHV7 A0A195FWP7 A0A195AUY0 A0A158NNA0 F4WLI7 E2A541 A0A3L8DGB5 A0A023FAA7 A0A224XQX1 A0A026WSS9 A0A069DTA0 A0A232EKU0 A0A1S4G7A8 A0A0J7KE25 A0A0N7Z9B7 A0A1Q3FTK8 A0A0M4EH57 E9IUE8 B4KKW7 A0A2W1BH33 A0A1W4WH07 K7JC85 A0A2M4CPV5 A0A2M4CPZ3 B4JQ84 A0A2R5L1M4 A0A0A1WJQ0 B4M9L1 A0A3Q0J971 B4N7N5 A0A2M4CPS9 A0A1B0Y0C3 W8BD39 A0A0P4W187 A0A0K8WKU8 A0A034VZ02 U5EG46 D6X452 A0A2M4BNS8 A0A2M4BSG5 A0A2J7RMR0 A0A1B6DXR3 A0A1B6J9K2 T1IAB8 B3N5S9 Q9VMC3 A0A1W4V270 B4P0C5 B4Q4X9 A0A3B0KBB5 A0A067RHX8 A0A0L0C491 A0A2M4BSU4 B3MP66 Q29PD2 A0A0T6AUP2 Q7Q1F9 A0A423SVV8 T1PJH1 A0A1S4H2X8 A0A182J7N5 A0A2C9GPU3 A0A084WG47 R4FKL8 A0A182V1S9 A0A1I8MIU1 A0A1D2MXQ9 E0VHS3 A0A1B6MIT2 A0A1I8Q9U4 W5J5F5 A0A182KTW2 A0A2R7WCE1 A0A1B6CQM9 A0A226EME3 A0A182Q212
A0A1E1W9V6 H9J4S9 A0A212F4E5 A0A154P009 A0A0M8ZY24 A0A0L7RAF8 A0A088ASZ0 A0A195CRL0 A0A151IT61 E2B3G5 A0A151XA79 A0A310SHV7 A0A195FWP7 A0A195AUY0 A0A158NNA0 F4WLI7 E2A541 A0A3L8DGB5 A0A023FAA7 A0A224XQX1 A0A026WSS9 A0A069DTA0 A0A232EKU0 A0A1S4G7A8 A0A0J7KE25 A0A0N7Z9B7 A0A1Q3FTK8 A0A0M4EH57 E9IUE8 B4KKW7 A0A2W1BH33 A0A1W4WH07 K7JC85 A0A2M4CPV5 A0A2M4CPZ3 B4JQ84 A0A2R5L1M4 A0A0A1WJQ0 B4M9L1 A0A3Q0J971 B4N7N5 A0A2M4CPS9 A0A1B0Y0C3 W8BD39 A0A0P4W187 A0A0K8WKU8 A0A034VZ02 U5EG46 D6X452 A0A2M4BNS8 A0A2M4BSG5 A0A2J7RMR0 A0A1B6DXR3 A0A1B6J9K2 T1IAB8 B3N5S9 Q9VMC3 A0A1W4V270 B4P0C5 B4Q4X9 A0A3B0KBB5 A0A067RHX8 A0A0L0C491 A0A2M4BSU4 B3MP66 Q29PD2 A0A0T6AUP2 Q7Q1F9 A0A423SVV8 T1PJH1 A0A1S4H2X8 A0A182J7N5 A0A2C9GPU3 A0A084WG47 R4FKL8 A0A182V1S9 A0A1I8MIU1 A0A1D2MXQ9 E0VHS3 A0A1B6MIT2 A0A1I8Q9U4 W5J5F5 A0A182KTW2 A0A2R7WCE1 A0A1B6CQM9 A0A226EME3 A0A182Q212
Ontologies
GO
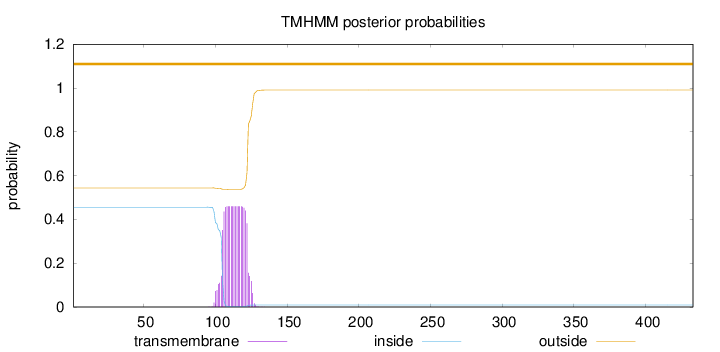
Topology
Length:
433
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
9.07757999999999
Exp number, first 60 AAs:
0.00082
Total prob of N-in:
0.45603
outside
1 - 433
Population Genetic Test Statistics
Pi
24.48138
Theta
185.023431
Tajima's D
-1.166827
CLR
1.115886
CSRT
0.112944352782361
Interpretation
Uncertain
