Gene
KWMTBOMO08840
Pre Gene Modal
BGIBMGA007854
Annotation
PREDICTED:_kynurenine_formamidase-like_isoform_X2_[Bombyx_mori]
Full name
Kynurenine formamidase
Alternative Name
Arylformamidase
N-formylkynurenine formamidase
N-formylkynurenine formamidase
Location in the cell
Nuclear Reliability : 1.41
Sequence
CDS
ATGGAATTTGTGGATTTGGAAAGAGAATACACACCGGTGTTGTGGTCGAACCGGTTATATACCCCGGAGCATCGCCTCAACTTGCACATCAACCTTGTAACAGCAGAGAGCGAAATAGCCACAAATAACGTTCCACACAAATTGGAGATTGAGTACGGATCAACGCCCGGACAGAAGCTGGACATTTTCGGTACCGACCTCCCGAATGAATCTCCAATTCTCGTGTTCATTCACGGTGGATACTGGCCGGACGTGTCTCGTGAAATATCACGTTATCCAGCAAAATCCCTCTACCCCGCCGGCGTTAAGACGATAATTGTTGGTTATGATCTCTGTCCCGCGGTCACGCTTGCTGAGGTCGTCAATCAGATACAGAACGCTGCGAGATACATCTTCGAATACGCTGAGAAGATGAATTCTAGAGGCGTTTACTTCGCGGGACACGGAACGGGAGCGCATCTGGTAGCGAAACTCCTGGCCAGTTCCGAATTCTTCGAAAACACGCCCGATTCACACCACCTCCAGGGAGCCTTCCTTATTTCAGGACTGTACGACTTACGGGAAGTCGTAAATACATCAGTCAACAGATCCGTGCAACTGCCTCACGAGTGGGCGGTGCCGCTGTCCCCGCAATTCGATTGTTTCTCCCACTTACAGTCGCGAAGGATCAGAATTTACATTTTTGCGGCCCAAAACGACAGCCCGACTTTCAAGAGGCAATCGAGAGAGTTCTATGAGCTTTTACACAATACGTGCTTGATGCAGAACATGTATTTGGAGATCAAAGACGATATGGATCATCACGAAATAGTCGAGTGCTTCGCGAACAGCAACAACTACCTTAAGAACTTGTTGATTCATGATATTCGTAAGCACATATGA
Protein
MEFVDLEREYTPVLWSNRLYTPEHRLNLHINLVTAESEIATNNVPHKLEIEYGSTPGQKLDIFGTDLPNESPILVFIHGGYWPDVSREISRYPAKSLYPAGVKTIIVGYDLCPAVTLAEVVNQIQNAARYIFEYAEKMNSRGVYFAGHGTGAHLVAKLLASSEFFENTPDSHHLQGAFLISGLYDLREVVNTSVNRSVQLPHEWAVPLSPQFDCFSHLQSRRIRIYIFAAQNDSPTFKRQSREFYELLHNTCLMQNMYLEIKDDMDHHEIVECFANSNNYLKNLLIHDIRKHI
Summary
Description
Catalyzes the hydrolysis of N-formyl-L-kynurenine to L-kynurenine, the second step in the kynurenine pathway of tryptophan degradation. Required for elimination of toxic metabolites.
Catalyzes the hydrolysis of N-formyl-L-kynurenine to L-kynurenine, the second step in the kynurenine pathway of tryptophan degradation. Kynurenine may be further oxidized to nicotinic acid, NAD(H) and NADP(H). Required for elimination of toxic metabolites.
Catalyzes the hydrolysis of N-formyl-L-kynurenine to L-kynurenine, the second step in the kynurenine pathway of tryptophan degradation. Kynurenine may be further oxidized to nicotinic acid, NAD(H) and NADP(H). Required for elimination of toxic metabolites.
Catalytic Activity
H2O + N-formyl-L-kynurenine = formate + H(+) + L-kynurenine
Subunit
Homodimer.
Similarity
Belongs to the kynurenine formamidase family.
Uniprot
H9JEA7
A0A194PQ23
A0A2A4JKB9
C6KEF1
A0A2A4JIQ8
A0A194QN01
+ More
A0A2W1B8N3 A0A212F2A7 A0A437AZ89 S4PP32 A0A0L7LV95 A0A2H1WNA8 H9JEA9 A0A1Y1MDP1 A0A067QLT8 U5ES04 A0A182QFV9 A0A182NEL5 A0A1W4XRI6 A0A1W4XFU9 A0A182G908 J3JUE8 U4U0B4 A0A182GZ03 A0A139WAT7 A0A1B6HEY5 A0A182PJ14 A0A182RG95 A0A182YFY2 Q1HRC1 V4ALI9 A0A1S4F7T3 Q0IFP1 A0A084VPT3 W5J900 A0A182VUS7 A0A190EFG9 B0W291 A0A182F5B4 Q7QKH2 A0A182MAI7 A0A182XIN4 U5EY00 A0A1S3KC92 A0A1S4H142 A0A182V8R0 A0A088A2A3 A0A1Q3FQI6 J3JZ36 A0A0P4ZP14 A0A1Y1MBD2 A0A182L5E7 A0A210QA33 A0A0P5FR89 A0A182HG53 A0A182J6H1 A0A232ELZ4 A0A182UBW5 A0A0P6B396 A0A1J1J319 A0A0L8GC40 A0A0P4XK47 E9GB07 A0A023F6Q9 A0A161M607 A0A2A3E9X7 A0A0P4VIP9 R4G805 A0A151XB44 A0A0P5C209 U4U343 A0A0J7P476 A0A224XKW5 A0A0P5XCD8 A0A195EQR9 A0A2R7WTW1 A0A0L8GC20 A0A1J1HHH1 N6TRP6 A0A0P5HH37 A0A1S3KCT5 A0A195DHX2 A0A195D3R2 A0A0P4ZJW9 A0A1B0CQ20 A0A0M9A453 A0A158NQK7 A0A0L7R3B9 E2BMI6 A0A0N8APU3 A0A182K2L6 F4WK97 A0A2J7QX32 E4WT97 H3BDX0 A0A210Q8U8 A0A1L8E2F9 E1ZXE8 A0A402F3Q9
A0A2W1B8N3 A0A212F2A7 A0A437AZ89 S4PP32 A0A0L7LV95 A0A2H1WNA8 H9JEA9 A0A1Y1MDP1 A0A067QLT8 U5ES04 A0A182QFV9 A0A182NEL5 A0A1W4XRI6 A0A1W4XFU9 A0A182G908 J3JUE8 U4U0B4 A0A182GZ03 A0A139WAT7 A0A1B6HEY5 A0A182PJ14 A0A182RG95 A0A182YFY2 Q1HRC1 V4ALI9 A0A1S4F7T3 Q0IFP1 A0A084VPT3 W5J900 A0A182VUS7 A0A190EFG9 B0W291 A0A182F5B4 Q7QKH2 A0A182MAI7 A0A182XIN4 U5EY00 A0A1S3KC92 A0A1S4H142 A0A182V8R0 A0A088A2A3 A0A1Q3FQI6 J3JZ36 A0A0P4ZP14 A0A1Y1MBD2 A0A182L5E7 A0A210QA33 A0A0P5FR89 A0A182HG53 A0A182J6H1 A0A232ELZ4 A0A182UBW5 A0A0P6B396 A0A1J1J319 A0A0L8GC40 A0A0P4XK47 E9GB07 A0A023F6Q9 A0A161M607 A0A2A3E9X7 A0A0P4VIP9 R4G805 A0A151XB44 A0A0P5C209 U4U343 A0A0J7P476 A0A224XKW5 A0A0P5XCD8 A0A195EQR9 A0A2R7WTW1 A0A0L8GC20 A0A1J1HHH1 N6TRP6 A0A0P5HH37 A0A1S3KCT5 A0A195DHX2 A0A195D3R2 A0A0P4ZJW9 A0A1B0CQ20 A0A0M9A453 A0A158NQK7 A0A0L7R3B9 E2BMI6 A0A0N8APU3 A0A182K2L6 F4WK97 A0A2J7QX32 E4WT97 H3BDX0 A0A210Q8U8 A0A1L8E2F9 E1ZXE8 A0A402F3Q9
EC Number
3.5.1.9
Pubmed
19121390
26354079
19754707
28756777
22118469
23622113
+ More
26227816 28004739 24845553 26483478 22516182 23537049 18362917 19820115 25244985 17204158 23254933 17510324 24438588 20920257 23761445 12364791 20966253 28812685 28648823 21292972 25474469 27129103 21347285 20798317 21719571 21097902 9215903
26227816 28004739 24845553 26483478 22516182 23537049 18362917 19820115 25244985 17204158 23254933 17510324 24438588 20920257 23761445 12364791 20966253 28812685 28648823 21292972 25474469 27129103 21347285 20798317 21719571 21097902 9215903
EMBL
BABH01002833
KQ459596
KPI95546.1
NWSH01001283
PCG71850.1
GQ183897
+ More
ACS66705.1 PCG71851.1 KQ461195 KPJ06734.1 KZ150220 PZC72109.1 AGBW02010781 OWR47862.1 RSAL01000249 RVE43512.1 GAIX01000467 JAA92093.1 JTDY01000021 KOB79382.1 ODYU01009469 SOQ53934.1 BABH01002838 BABH01002839 BABH01002840 GEZM01034617 JAV83701.1 KK853274 KDR09098.1 GANO01002587 JAB57284.1 AXCN02000850 JXUM01048555 KQ561567 KXJ78141.1 BT126861 AEE61823.1 KB630055 KB630056 KB630208 ERL83365.1 ERL83367.1 ERL83475.1 JXUM01098528 KQ564427 KXJ72302.1 KQ971380 KYB25017.1 GECU01034501 JAS73205.1 DQ440173 ABF18206.1 KB199651 ESP05049.1 CH477306 EAT44101.1 ATLV01015034 KE524999 KFB39977.1 ADMH02002093 ETN59279.1 KP881330 ALQ52681.1 DS231825 EDS28155.1 AAAB01008797 EAA03628.4 AXCM01004322 GANO01002267 JAB57604.1 GFDL01005263 JAV29782.1 BT128517 AEE63474.1 GDIP01214349 LRGB01002076 JAJ09053.1 KZS09367.1 GEZM01035656 JAV83139.1 NEDP02004470 OWF45559.1 GDIQ01259020 JAJ92704.1 APCN01004115 NNAY01003463 OXU19366.1 GDIP01034395 JAM69320.1 CVRI01000067 CRL06829.1 KQ422595 KOF74596.1 GDIP01249797 JAI73604.1 GL732537 EFX83454.1 GBBI01001666 JAC17046.1 GEMB01001942 JAS01233.1 KZ288311 PBC28520.1 GDKW01001846 JAI54749.1 ACPB03019872 GAHY01001812 JAA75698.1 KQ982335 KYQ57520.1 GDIP01176804 JAJ46598.1 KB631855 ERL86758.1 LBMM01000146 KMR04739.1 GFTR01004712 JAW11714.1 GDIP01074172 JAM29543.1 KQ982021 KYN30558.1 KK855326 PTY22025.1 KOF74597.1 CVRI01000004 CRK87453.1 APGK01057597 KB741280 ENN70981.1 GDIQ01228116 JAK23609.1 KQ980824 KYN12495.1 KQ976885 KYN07538.1 GDIP01211702 JAJ11700.1 AJWK01022926 KQ435762 KOX75479.1 ADTU01023371 KQ414663 KOC65362.1 GL449233 EFN83095.1 GDIQ01254715 JAJ97009.1 GL888199 EGI65321.1 NEVH01009397 PNF33146.1 FN653016 CBY06998.1 AFYH01017047 NEDP02004580 OWF45109.1 GFDF01001187 JAV12897.1 GL435038 EFN74125.1 BDOT01000098 GCF51165.1
ACS66705.1 PCG71851.1 KQ461195 KPJ06734.1 KZ150220 PZC72109.1 AGBW02010781 OWR47862.1 RSAL01000249 RVE43512.1 GAIX01000467 JAA92093.1 JTDY01000021 KOB79382.1 ODYU01009469 SOQ53934.1 BABH01002838 BABH01002839 BABH01002840 GEZM01034617 JAV83701.1 KK853274 KDR09098.1 GANO01002587 JAB57284.1 AXCN02000850 JXUM01048555 KQ561567 KXJ78141.1 BT126861 AEE61823.1 KB630055 KB630056 KB630208 ERL83365.1 ERL83367.1 ERL83475.1 JXUM01098528 KQ564427 KXJ72302.1 KQ971380 KYB25017.1 GECU01034501 JAS73205.1 DQ440173 ABF18206.1 KB199651 ESP05049.1 CH477306 EAT44101.1 ATLV01015034 KE524999 KFB39977.1 ADMH02002093 ETN59279.1 KP881330 ALQ52681.1 DS231825 EDS28155.1 AAAB01008797 EAA03628.4 AXCM01004322 GANO01002267 JAB57604.1 GFDL01005263 JAV29782.1 BT128517 AEE63474.1 GDIP01214349 LRGB01002076 JAJ09053.1 KZS09367.1 GEZM01035656 JAV83139.1 NEDP02004470 OWF45559.1 GDIQ01259020 JAJ92704.1 APCN01004115 NNAY01003463 OXU19366.1 GDIP01034395 JAM69320.1 CVRI01000067 CRL06829.1 KQ422595 KOF74596.1 GDIP01249797 JAI73604.1 GL732537 EFX83454.1 GBBI01001666 JAC17046.1 GEMB01001942 JAS01233.1 KZ288311 PBC28520.1 GDKW01001846 JAI54749.1 ACPB03019872 GAHY01001812 JAA75698.1 KQ982335 KYQ57520.1 GDIP01176804 JAJ46598.1 KB631855 ERL86758.1 LBMM01000146 KMR04739.1 GFTR01004712 JAW11714.1 GDIP01074172 JAM29543.1 KQ982021 KYN30558.1 KK855326 PTY22025.1 KOF74597.1 CVRI01000004 CRK87453.1 APGK01057597 KB741280 ENN70981.1 GDIQ01228116 JAK23609.1 KQ980824 KYN12495.1 KQ976885 KYN07538.1 GDIP01211702 JAJ11700.1 AJWK01022926 KQ435762 KOX75479.1 ADTU01023371 KQ414663 KOC65362.1 GL449233 EFN83095.1 GDIQ01254715 JAJ97009.1 GL888199 EGI65321.1 NEVH01009397 PNF33146.1 FN653016 CBY06998.1 AFYH01017047 NEDP02004580 OWF45109.1 GFDF01001187 JAV12897.1 GL435038 EFN74125.1 BDOT01000098 GCF51165.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000053240
UP000007151
UP000283053
+ More
UP000037510 UP000027135 UP000075886 UP000075884 UP000192223 UP000069940 UP000249989 UP000030742 UP000007266 UP000075885 UP000075900 UP000076408 UP000030746 UP000008820 UP000030765 UP000000673 UP000075920 UP000002320 UP000069272 UP000007062 UP000075883 UP000076407 UP000085678 UP000075903 UP000005203 UP000076858 UP000075882 UP000242188 UP000075840 UP000075880 UP000215335 UP000075902 UP000183832 UP000053454 UP000000305 UP000242457 UP000015103 UP000075809 UP000036403 UP000078541 UP000019118 UP000078492 UP000078542 UP000092461 UP000053105 UP000005205 UP000053825 UP000008237 UP000075881 UP000007755 UP000235965 UP000008672 UP000000311 UP000288954
UP000037510 UP000027135 UP000075886 UP000075884 UP000192223 UP000069940 UP000249989 UP000030742 UP000007266 UP000075885 UP000075900 UP000076408 UP000030746 UP000008820 UP000030765 UP000000673 UP000075920 UP000002320 UP000069272 UP000007062 UP000075883 UP000076407 UP000085678 UP000075903 UP000005203 UP000076858 UP000075882 UP000242188 UP000075840 UP000075880 UP000215335 UP000075902 UP000183832 UP000053454 UP000000305 UP000242457 UP000015103 UP000075809 UP000036403 UP000078541 UP000019118 UP000078492 UP000078542 UP000092461 UP000053105 UP000005205 UP000053825 UP000008237 UP000075881 UP000007755 UP000235965 UP000008672 UP000000311 UP000288954
Interpro
Gene 3D
ProteinModelPortal
H9JEA7
A0A194PQ23
A0A2A4JKB9
C6KEF1
A0A2A4JIQ8
A0A194QN01
+ More
A0A2W1B8N3 A0A212F2A7 A0A437AZ89 S4PP32 A0A0L7LV95 A0A2H1WNA8 H9JEA9 A0A1Y1MDP1 A0A067QLT8 U5ES04 A0A182QFV9 A0A182NEL5 A0A1W4XRI6 A0A1W4XFU9 A0A182G908 J3JUE8 U4U0B4 A0A182GZ03 A0A139WAT7 A0A1B6HEY5 A0A182PJ14 A0A182RG95 A0A182YFY2 Q1HRC1 V4ALI9 A0A1S4F7T3 Q0IFP1 A0A084VPT3 W5J900 A0A182VUS7 A0A190EFG9 B0W291 A0A182F5B4 Q7QKH2 A0A182MAI7 A0A182XIN4 U5EY00 A0A1S3KC92 A0A1S4H142 A0A182V8R0 A0A088A2A3 A0A1Q3FQI6 J3JZ36 A0A0P4ZP14 A0A1Y1MBD2 A0A182L5E7 A0A210QA33 A0A0P5FR89 A0A182HG53 A0A182J6H1 A0A232ELZ4 A0A182UBW5 A0A0P6B396 A0A1J1J319 A0A0L8GC40 A0A0P4XK47 E9GB07 A0A023F6Q9 A0A161M607 A0A2A3E9X7 A0A0P4VIP9 R4G805 A0A151XB44 A0A0P5C209 U4U343 A0A0J7P476 A0A224XKW5 A0A0P5XCD8 A0A195EQR9 A0A2R7WTW1 A0A0L8GC20 A0A1J1HHH1 N6TRP6 A0A0P5HH37 A0A1S3KCT5 A0A195DHX2 A0A195D3R2 A0A0P4ZJW9 A0A1B0CQ20 A0A0M9A453 A0A158NQK7 A0A0L7R3B9 E2BMI6 A0A0N8APU3 A0A182K2L6 F4WK97 A0A2J7QX32 E4WT97 H3BDX0 A0A210Q8U8 A0A1L8E2F9 E1ZXE8 A0A402F3Q9
A0A2W1B8N3 A0A212F2A7 A0A437AZ89 S4PP32 A0A0L7LV95 A0A2H1WNA8 H9JEA9 A0A1Y1MDP1 A0A067QLT8 U5ES04 A0A182QFV9 A0A182NEL5 A0A1W4XRI6 A0A1W4XFU9 A0A182G908 J3JUE8 U4U0B4 A0A182GZ03 A0A139WAT7 A0A1B6HEY5 A0A182PJ14 A0A182RG95 A0A182YFY2 Q1HRC1 V4ALI9 A0A1S4F7T3 Q0IFP1 A0A084VPT3 W5J900 A0A182VUS7 A0A190EFG9 B0W291 A0A182F5B4 Q7QKH2 A0A182MAI7 A0A182XIN4 U5EY00 A0A1S3KC92 A0A1S4H142 A0A182V8R0 A0A088A2A3 A0A1Q3FQI6 J3JZ36 A0A0P4ZP14 A0A1Y1MBD2 A0A182L5E7 A0A210QA33 A0A0P5FR89 A0A182HG53 A0A182J6H1 A0A232ELZ4 A0A182UBW5 A0A0P6B396 A0A1J1J319 A0A0L8GC40 A0A0P4XK47 E9GB07 A0A023F6Q9 A0A161M607 A0A2A3E9X7 A0A0P4VIP9 R4G805 A0A151XB44 A0A0P5C209 U4U343 A0A0J7P476 A0A224XKW5 A0A0P5XCD8 A0A195EQR9 A0A2R7WTW1 A0A0L8GC20 A0A1J1HHH1 N6TRP6 A0A0P5HH37 A0A1S3KCT5 A0A195DHX2 A0A195D3R2 A0A0P4ZJW9 A0A1B0CQ20 A0A0M9A453 A0A158NQK7 A0A0L7R3B9 E2BMI6 A0A0N8APU3 A0A182K2L6 F4WK97 A0A2J7QX32 E4WT97 H3BDX0 A0A210Q8U8 A0A1L8E2F9 E1ZXE8 A0A402F3Q9
PDB
4E15
E-value=4.08913e-23,
Score=266
Ontologies
PATHWAY
GO
Topology
Subcellular location
Cytoplasm
Cytosol
Nucleus
Cytosol
Nucleus
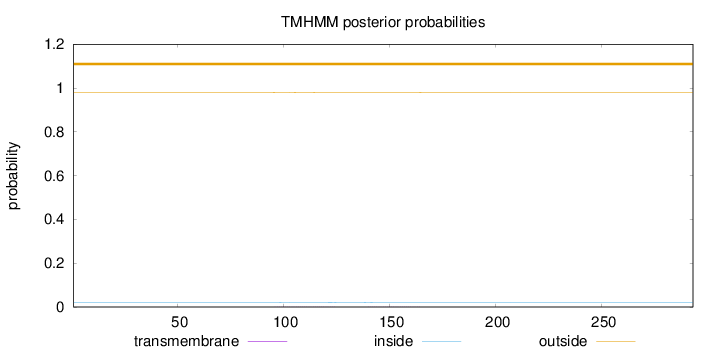
Length:
293
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0361600000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01997
outside
1 - 293
Population Genetic Test Statistics
Pi
197.697802
Theta
156.940352
Tajima's D
0.80643
CLR
0.271407
CSRT
0.612669366531673
Interpretation
Uncertain
