Gene
KWMTBOMO08820 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007701
Annotation
abnormal_wing_disc-like_protein_[Bombyx_mori]
Full name
Nucleoside diphosphate kinase
Alternative Name
Abnormal wing disks protein
Killer of prune protein
Killer of prune protein
Location in the cell
Cytoplasmic Reliability : 2.228
Sequence
CDS
ATGGCGGAACAACGTGAGCGTACTTTTATTATGGTTAAGCCTGATGGTGTACAACGTGGTCTTGTGGGCACCATTATTGAACGTTTCGAAAAGAAAGGCTTCAAACTAGTCGGTTTGAAATTCGTATGGCCATCAGAAGAACTTCTCCAGCAACACTACAGCGATTTGGCATCCCGGCCTTTCTTCCCTGGTCTAGTAAAGTACATGAGTTCAGGACCTGTGGTCCCTATGGTATGGGAGGGCCTTAATGTTGTGAAGACTGGCCGTCAAATGCTTGGCGCAACTAACCCAGCTGACTCGCAGCCCGGCACTATCCGCGGTGATCTCTGCATTCAAGTTGGGCGTAACATCATCCATGGTTCGGACAGCGTTGAATCTGCTAAAAAGGAAATAGGCCTCTGGTTTACGGACAAAGAAGTTGTGGGCTGGACACCTGCAAATGAAAACTGGGTTTATGAGTAA
Protein
MAEQRERTFIMVKPDGVQRGLVGTIIERFEKKGFKLVGLKFVWPSEELLQQHYSDLASRPFFPGLVKYMSSGPVVPMVWEGLNVVKTGRQMLGATNPADSQPGTIRGDLCIQVGRNIIHGSDSVESAKKEIGLWFTDKEVVGWTPANENWVYE
Summary
Description
Major role in the synthesis of nucleoside triphosphates other than ATP. The ATP gamma phosphate is transferred to the NDP beta phosphate via a ping-pong mechanism, using a phosphorylated active-site intermediate.
Major role in the synthesis of nucleoside triphosphates other than ATP. The ATP gamma phosphate is transferred to the NDP beta phosphate via a ping-pong mechanism, using a phosphorylated active-site intermediate (By similarity).
Major role in the synthesis of nucleoside triphosphates other than ATP. The ATP gamma phosphate is transferred to the NDP beta phosphate via a ping-pong mechanism, using a phosphorylated active-site intermediate (By similarity).
Catalytic Activity
a 2'-deoxyribonucleoside 5'-diphosphate + ATP = a 2'-deoxyribonucleoside 5'-triphosphate + ADP
a ribonucleoside 5'-diphosphate + ATP = a ribonucleoside 5'-triphosphate + ADP
a ribonucleoside 5'-diphosphate + ATP = a ribonucleoside 5'-triphosphate + ADP
Cofactor
Mg(2+)
Subunit
Homohexamer.
Miscellaneous
Mutations cause abnormal tissue morphology and necrosis and widespread aberrant differentiation.
Similarity
Belongs to the NDK family.
Keywords
3D-structure
Acetylation
ATP-binding
Complete proteome
Cytoplasm
Cytoskeleton
Direct protein sequencing
Kinase
Magnesium
Metal-binding
Microtubule
Nucleotide metabolism
Nucleotide-binding
Phosphoprotein
Reference proteome
Transferase
Feature
chain Nucleoside diphosphate kinase
Uniprot
Q1HPI5
H9JDV4
A0A3S2NSB7
E3SRH2
A0A170QHP8
I4DJ21
+ More
A0A194QMH0 A0A194PS06 A0A2A4JIZ8 A0A2H1VK95 A0A2W1BB94 I4DML6 H8YI03 S4P4M4 A0A3G1T1K4 A0A212FEK6 Q8MUR5 A0A0L7KXM5 D6WHK2 A0A0T6B1P4 A0A0A7BYH5 A0A1S3DHQ1 Q0PXX9 U5ELM6 A0A2M4A0G9 A0A2M4A0J5 A0A1B0D3Q5 A0A182WLI1 T1EA13 A0A182F9R5 A0A182YEK8 A0A182QJ33 A0A182P0N1 A0A182M9A4 K7QJQ7 A0A1Q3FWR1 A0A2M4C1E2 A0A2M4C1H6 A0A2M4AWP1 B6DDT8 B3MSX7 W5JV72 A0A182NK61 A0A182V070 A0A182UBS7 A0A182WWY7 A0A182KQD4 Q7QIX6 A0A182I2I6 A0A182JND1 A0A2M4CI12 A8E6J4 B4PP18 B3P511 B4N8T2 B4QT92 A0A084VDH9 J3JZ25 A0A182RP58 N6UB03 A0A1L8DKD9 A0A3B0JL30 A0A0B4LHX6 B4KCG6 W8C3P8 T1DIW5 B4IJ26 A0A0B4LJ12 P08879 A0A0J7KSF4 A0A026WVX5 A0A1W4W6Y4 B5DW72 A0A2M3YYQ7 A0A067QR89 A0A0C9RVE7 A0A034WTB1 A0A182IPA3 R4UKI3 A0A0K8VUG3 B4M0X8 B4G2Y5 A0A023EHG0 A0A0P6IJK1 Q16MB4 A0A023EHV4 E9IUS3 E2AQV0 A0A1L8EIF7 A0A1W4WP60 A0A182GKP0 A0A1L8EII8 B4JH64 A0A1L8EIA0 A0A1D1YPU1 A0A151WHF8 Q6XI71 A0A1I8M733 A0A151JSH4 A0A195F0E8
A0A194QMH0 A0A194PS06 A0A2A4JIZ8 A0A2H1VK95 A0A2W1BB94 I4DML6 H8YI03 S4P4M4 A0A3G1T1K4 A0A212FEK6 Q8MUR5 A0A0L7KXM5 D6WHK2 A0A0T6B1P4 A0A0A7BYH5 A0A1S3DHQ1 Q0PXX9 U5ELM6 A0A2M4A0G9 A0A2M4A0J5 A0A1B0D3Q5 A0A182WLI1 T1EA13 A0A182F9R5 A0A182YEK8 A0A182QJ33 A0A182P0N1 A0A182M9A4 K7QJQ7 A0A1Q3FWR1 A0A2M4C1E2 A0A2M4C1H6 A0A2M4AWP1 B6DDT8 B3MSX7 W5JV72 A0A182NK61 A0A182V070 A0A182UBS7 A0A182WWY7 A0A182KQD4 Q7QIX6 A0A182I2I6 A0A182JND1 A0A2M4CI12 A8E6J4 B4PP18 B3P511 B4N8T2 B4QT92 A0A084VDH9 J3JZ25 A0A182RP58 N6UB03 A0A1L8DKD9 A0A3B0JL30 A0A0B4LHX6 B4KCG6 W8C3P8 T1DIW5 B4IJ26 A0A0B4LJ12 P08879 A0A0J7KSF4 A0A026WVX5 A0A1W4W6Y4 B5DW72 A0A2M3YYQ7 A0A067QR89 A0A0C9RVE7 A0A034WTB1 A0A182IPA3 R4UKI3 A0A0K8VUG3 B4M0X8 B4G2Y5 A0A023EHG0 A0A0P6IJK1 Q16MB4 A0A023EHV4 E9IUS3 E2AQV0 A0A1L8EIF7 A0A1W4WP60 A0A182GKP0 A0A1L8EII8 B4JH64 A0A1L8EIA0 A0A1D1YPU1 A0A151WHF8 Q6XI71 A0A1I8M733 A0A151JSH4 A0A195F0E8
EC Number
2.7.4.6
Pubmed
19121390
22651552
26354079
28756777
23468937
23622113
+ More
22118469 26227816 18362917 19820115 25244985 23126268 19178717 17994087 20920257 23761445 20966253 12364791 14747013 17210077 17550304 24438588 22516182 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 24495485 24330624 26109357 26109356 2849580 12537569 12099695 8500545 2175255 1320004 7559441 18327897 8081741 7669763 24508170 15632085 24845553 25348373 24945155 17510324 26483478 21282665 20798317 14525923 25315136
22118469 26227816 18362917 19820115 25244985 23126268 19178717 17994087 20920257 23761445 20966253 12364791 14747013 17210077 17550304 24438588 22516182 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 24495485 24330624 26109357 26109356 2849580 12537569 12099695 8500545 2175255 1320004 7559441 18327897 8081741 7669763 24508170 15632085 24845553 25348373 24945155 17510324 26483478 21282665 20798317 14525923 25315136
EMBL
DQ443417
ABF51506.1
BABH01002865
RSAL01000266
RVE43271.1
GU073317
+ More
ADE75591.1 KT984956 AMY63175.1 AK401289 BAM17911.1 KQ461195 KPJ06722.1 KQ459596 KPI95534.1 NWSH01001323 PCG71688.1 ODYU01003012 SOQ41243.1 KZ150372 PZC71134.1 AK402534 BAM19156.1 JF690666 AEC12435.1 GAIX01005904 JAA86656.1 MG992433 AXY94871.1 AGBW02008937 OWR52127.1 AF514332 AAM53644.1 JTDY01004756 KOB67796.1 KQ971331 EEZ99726.1 LJIG01016195 KRT81331.1 KC966938 AHV85212.1 DQ673407 ABG81980.1 GANO01001340 JAB58531.1 GGFK01000983 MBW34304.1 GGFK01000982 MBW34303.1 AJVK01011006 GAMD01001428 JAB00163.1 AXCN02000429 AXCM01001237 JQ085374 JQ085375 AFU52975.1 GFDL01003026 JAV32019.1 GGFJ01009982 MBW59123.1 GGFJ01010016 MBW59157.1 GGFK01011895 MBW45216.1 EU934298 ACI30076.1 CH902623 EDV30367.2 ADMH02000262 ETN67208.1 AAAB01008807 EAA04524.4 APCN01000164 GGFL01000812 MBW64990.1 BT030786 ABV82168.1 CM000160 EDW99254.1 CH954182 EDV52925.1 CH964232 EDW81533.2 CM000364 EDX15154.1 ATLV01011337 KE524664 KFB36023.1 BT128506 AEE63463.1 APGK01041853 KB740998 KB632355 ENN75817.1 ERL93384.1 GFDF01007156 JAV06928.1 OUUW01000005 SPP81032.1 AE014297 AHN57624.1 CH933806 EDW14785.2 GAMC01005979 JAC00577.1 GALA01000706 JAA94146.1 CH480846 EDW50979.1 KX531763 AHN57623.1 ANY27573.1 X13107 AY113576 LBMM01003772 KMQ93159.1 KK107078 EZA60210.1 CM000070 EDY68117.2 GGFM01000610 MBW21361.1 KK853114 KDR11183.1 GBYB01012880 JAG82647.1 GAKP01001380 JAC57572.1 KC740805 AGM32629.1 GDHF01009787 JAI42527.1 CH940650 EDW67389.2 CH479179 EDW24180.1 GAPW01005141 JAC08457.1 GDIQ01003407 JAN91330.1 CH477869 EAT35464.1 JXUM01024999 GAPW01005142 KQ560708 JAC08456.1 KXJ81204.1 GL766032 EFZ15678.1 GL441834 EFN64212.1 GFDG01000364 JAV18435.1 JXUM01071189 KQ562648 KXJ75411.1 GFDG01000363 JAV18436.1 CH916369 EDV92755.1 GFDG01000362 JAV18437.1 GDJX01011285 JAT56651.1 KQ983120 KYQ47247.1 AY231961 AAR09984.1 KQ978530 KYN30292.1 KQ981866 KYN34055.1
ADE75591.1 KT984956 AMY63175.1 AK401289 BAM17911.1 KQ461195 KPJ06722.1 KQ459596 KPI95534.1 NWSH01001323 PCG71688.1 ODYU01003012 SOQ41243.1 KZ150372 PZC71134.1 AK402534 BAM19156.1 JF690666 AEC12435.1 GAIX01005904 JAA86656.1 MG992433 AXY94871.1 AGBW02008937 OWR52127.1 AF514332 AAM53644.1 JTDY01004756 KOB67796.1 KQ971331 EEZ99726.1 LJIG01016195 KRT81331.1 KC966938 AHV85212.1 DQ673407 ABG81980.1 GANO01001340 JAB58531.1 GGFK01000983 MBW34304.1 GGFK01000982 MBW34303.1 AJVK01011006 GAMD01001428 JAB00163.1 AXCN02000429 AXCM01001237 JQ085374 JQ085375 AFU52975.1 GFDL01003026 JAV32019.1 GGFJ01009982 MBW59123.1 GGFJ01010016 MBW59157.1 GGFK01011895 MBW45216.1 EU934298 ACI30076.1 CH902623 EDV30367.2 ADMH02000262 ETN67208.1 AAAB01008807 EAA04524.4 APCN01000164 GGFL01000812 MBW64990.1 BT030786 ABV82168.1 CM000160 EDW99254.1 CH954182 EDV52925.1 CH964232 EDW81533.2 CM000364 EDX15154.1 ATLV01011337 KE524664 KFB36023.1 BT128506 AEE63463.1 APGK01041853 KB740998 KB632355 ENN75817.1 ERL93384.1 GFDF01007156 JAV06928.1 OUUW01000005 SPP81032.1 AE014297 AHN57624.1 CH933806 EDW14785.2 GAMC01005979 JAC00577.1 GALA01000706 JAA94146.1 CH480846 EDW50979.1 KX531763 AHN57623.1 ANY27573.1 X13107 AY113576 LBMM01003772 KMQ93159.1 KK107078 EZA60210.1 CM000070 EDY68117.2 GGFM01000610 MBW21361.1 KK853114 KDR11183.1 GBYB01012880 JAG82647.1 GAKP01001380 JAC57572.1 KC740805 AGM32629.1 GDHF01009787 JAI42527.1 CH940650 EDW67389.2 CH479179 EDW24180.1 GAPW01005141 JAC08457.1 GDIQ01003407 JAN91330.1 CH477869 EAT35464.1 JXUM01024999 GAPW01005142 KQ560708 JAC08456.1 KXJ81204.1 GL766032 EFZ15678.1 GL441834 EFN64212.1 GFDG01000364 JAV18435.1 JXUM01071189 KQ562648 KXJ75411.1 GFDG01000363 JAV18436.1 CH916369 EDV92755.1 GFDG01000362 JAV18437.1 GDJX01011285 JAT56651.1 KQ983120 KYQ47247.1 AY231961 AAR09984.1 KQ978530 KYN30292.1 KQ981866 KYN34055.1
Proteomes
UP000005204
UP000283053
UP000053240
UP000053268
UP000218220
UP000007151
+ More
UP000037510 UP000007266 UP000079169 UP000092462 UP000075920 UP000069272 UP000076408 UP000075886 UP000075885 UP000075883 UP000007801 UP000000673 UP000075884 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000075881 UP000002282 UP000008711 UP000007798 UP000000304 UP000030765 UP000075900 UP000019118 UP000030742 UP000268350 UP000000803 UP000009192 UP000001292 UP000036403 UP000053097 UP000192221 UP000001819 UP000027135 UP000075880 UP000008792 UP000008744 UP000008820 UP000069940 UP000249989 UP000000311 UP000192223 UP000001070 UP000075809 UP000095301 UP000078492 UP000078541
UP000037510 UP000007266 UP000079169 UP000092462 UP000075920 UP000069272 UP000076408 UP000075886 UP000075885 UP000075883 UP000007801 UP000000673 UP000075884 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000075881 UP000002282 UP000008711 UP000007798 UP000000304 UP000030765 UP000075900 UP000019118 UP000030742 UP000268350 UP000000803 UP000009192 UP000001292 UP000036403 UP000053097 UP000192221 UP000001819 UP000027135 UP000075880 UP000008792 UP000008744 UP000008820 UP000069940 UP000249989 UP000000311 UP000192223 UP000001070 UP000075809 UP000095301 UP000078492 UP000078541
Pfam
PF00334 NDK
Interpro
SUPFAM
SSF54919
SSF54919
Gene 3D
ProteinModelPortal
Q1HPI5
H9JDV4
A0A3S2NSB7
E3SRH2
A0A170QHP8
I4DJ21
+ More
A0A194QMH0 A0A194PS06 A0A2A4JIZ8 A0A2H1VK95 A0A2W1BB94 I4DML6 H8YI03 S4P4M4 A0A3G1T1K4 A0A212FEK6 Q8MUR5 A0A0L7KXM5 D6WHK2 A0A0T6B1P4 A0A0A7BYH5 A0A1S3DHQ1 Q0PXX9 U5ELM6 A0A2M4A0G9 A0A2M4A0J5 A0A1B0D3Q5 A0A182WLI1 T1EA13 A0A182F9R5 A0A182YEK8 A0A182QJ33 A0A182P0N1 A0A182M9A4 K7QJQ7 A0A1Q3FWR1 A0A2M4C1E2 A0A2M4C1H6 A0A2M4AWP1 B6DDT8 B3MSX7 W5JV72 A0A182NK61 A0A182V070 A0A182UBS7 A0A182WWY7 A0A182KQD4 Q7QIX6 A0A182I2I6 A0A182JND1 A0A2M4CI12 A8E6J4 B4PP18 B3P511 B4N8T2 B4QT92 A0A084VDH9 J3JZ25 A0A182RP58 N6UB03 A0A1L8DKD9 A0A3B0JL30 A0A0B4LHX6 B4KCG6 W8C3P8 T1DIW5 B4IJ26 A0A0B4LJ12 P08879 A0A0J7KSF4 A0A026WVX5 A0A1W4W6Y4 B5DW72 A0A2M3YYQ7 A0A067QR89 A0A0C9RVE7 A0A034WTB1 A0A182IPA3 R4UKI3 A0A0K8VUG3 B4M0X8 B4G2Y5 A0A023EHG0 A0A0P6IJK1 Q16MB4 A0A023EHV4 E9IUS3 E2AQV0 A0A1L8EIF7 A0A1W4WP60 A0A182GKP0 A0A1L8EII8 B4JH64 A0A1L8EIA0 A0A1D1YPU1 A0A151WHF8 Q6XI71 A0A1I8M733 A0A151JSH4 A0A195F0E8
A0A194QMH0 A0A194PS06 A0A2A4JIZ8 A0A2H1VK95 A0A2W1BB94 I4DML6 H8YI03 S4P4M4 A0A3G1T1K4 A0A212FEK6 Q8MUR5 A0A0L7KXM5 D6WHK2 A0A0T6B1P4 A0A0A7BYH5 A0A1S3DHQ1 Q0PXX9 U5ELM6 A0A2M4A0G9 A0A2M4A0J5 A0A1B0D3Q5 A0A182WLI1 T1EA13 A0A182F9R5 A0A182YEK8 A0A182QJ33 A0A182P0N1 A0A182M9A4 K7QJQ7 A0A1Q3FWR1 A0A2M4C1E2 A0A2M4C1H6 A0A2M4AWP1 B6DDT8 B3MSX7 W5JV72 A0A182NK61 A0A182V070 A0A182UBS7 A0A182WWY7 A0A182KQD4 Q7QIX6 A0A182I2I6 A0A182JND1 A0A2M4CI12 A8E6J4 B4PP18 B3P511 B4N8T2 B4QT92 A0A084VDH9 J3JZ25 A0A182RP58 N6UB03 A0A1L8DKD9 A0A3B0JL30 A0A0B4LHX6 B4KCG6 W8C3P8 T1DIW5 B4IJ26 A0A0B4LJ12 P08879 A0A0J7KSF4 A0A026WVX5 A0A1W4W6Y4 B5DW72 A0A2M3YYQ7 A0A067QR89 A0A0C9RVE7 A0A034WTB1 A0A182IPA3 R4UKI3 A0A0K8VUG3 B4M0X8 B4G2Y5 A0A023EHG0 A0A0P6IJK1 Q16MB4 A0A023EHV4 E9IUS3 E2AQV0 A0A1L8EIF7 A0A1W4WP60 A0A182GKP0 A0A1L8EII8 B4JH64 A0A1L8EIA0 A0A1D1YPU1 A0A151WHF8 Q6XI71 A0A1I8M733 A0A151JSH4 A0A195F0E8
PDB
1NSQ
E-value=7.55304e-72,
Score=682
Ontologies
PATHWAY
GO
GO:0004550
GO:0006183
GO:0005524
GO:0006228
GO:0006241
GO:0004519
GO:0006468
GO:0005737
GO:0009117
GO:0007424
GO:0018105
GO:0006165
GO:0000287
GO:0016301
GO:0046777
GO:0005886
GO:0007017
GO:0007427
GO:0005880
GO:0005874
GO:0008017
GO:0000278
GO:0005525
GO:0035152
GO:0034332
GO:0016334
GO:0016021
GO:0016817
GO:0008270
GO:0043565
GO:0016876
GO:0043039
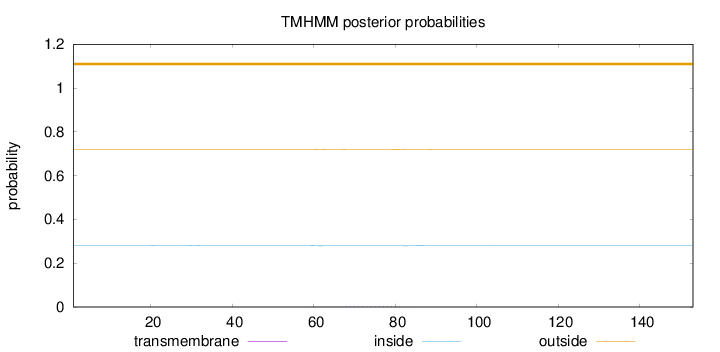
Topology
Subcellular location
Cytoplasm
Cytoskeleton
Cytoskeleton
Length:
153
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.05455
Exp number, first 60 AAs:
0.0018
Total prob of N-in:
0.28055
outside
1 - 153
Population Genetic Test Statistics
Pi
192.949459
Theta
152.511759
Tajima's D
0.782539
CLR
0
CSRT
0.598270086495675
Interpretation
Uncertain
