Gene
KWMTBOMO08815 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007869
Annotation
PREDICTED:_ATP-binding_cassette_sub-family_F_member_1_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 4.312
Sequence
CDS
ATGTCGAAAAAGAAAGGAAACAAAAAAAATCAAGATTTAGATGACGATTTCGATGAAAAGCCTTCAGTCGTGAATGAAAAGACAGAACTAACCTCTAAAAATAAAGGAAAAGGCAAGAAGAAGAATAAAACTGGAGATTGTAGCGATAGCGACGAAGAGAACATAAAGAAAGTTCCAGTTTCAGACGAAGAGGACGTCCCGAAACCGGCTGCCAAAAAGTCACAAAAGAAAGGAAAGAGTAAAAAAAAGGGTGCTGATAGGTCAGATGATGAAGAGGATGATAACAAATCCATTATCAACAATACAACAAATGTTTCATCAAAGCCAGCTAAACTTGCAAAAAAGAAAAAAGGGAAAGGTAAGAAAGATGATGATTGGTCTTCGGAAGGCAGTGATGTAGATCTCAAAATGGCTTCAGATGAAGATGTTGCAAAGCCTGTTGGTAAGAAGAAAACCAAGGGTAAAAAGGATGACTTACCTGATGAAGACAGTGACAAAGAATTGAAAATTGAAGCTTCAGAAGATGATGAATTAGTCATACCTTTAGTAAAAAGTAAAGGAAAACATAAAAAGAAGAATAATATGGATGATATTGAGGCAGAGTTAAAGAAATTTGAAATAAAAGATCCAGAGCATGAAGAGAAATCGCTAAAGGGCAAAGATAAAAAGAAAAAGCGACAAGAAGATGAATCCATTGTTGAAGAAAAAGATGACGATAAGGTGACAGAATTGCCTTCAAAAGAAAAAGAAGATTCTGTAGAAAAGAAATTGAGCCACAAAGAAAAGAAGAAACTGAAGAAACAGCAGGAGTATGAGAAACAGATGGAATTGATGACAAAAAAAGGTGGTCAGGGGCACAGTGAACTTGATGCAAACTTCACGGTCAGTCAGGCACAGAAAACAGCTGGTCAAATGGCAGCCCTAGAAAACGCAGTAGACATAAAAGTCGAAAACTTCTCGATCAGTGCGAAAGGAAAAGATTTATTCTTGAACGCGAATCTCTTGATTGCCAACGGACGTCGATATGGTTTGGTCGGGCCTAATGGACACGGAAAAACAACTTTATTGAGGCATTTAGCACAACGCGCCTTCCCGCTCCCGCCCCATATAGACATACTGTTGTGCGAGCAAGAGGTGACGGCCAGTGATATGTCAGCTGTAGATACGCTCCTCGAATCTGATGTTAAGAGAACCGAATTACTAAAGGAATGCAAAGAGCTTGAAGCTGAAACAGAGAAAGGCAACCTCAGCAAACAGGATAGACTTAATGAGGTTTACGCAGAACTGAAAGCGATAGGAGCGGACGCGGCTGAGCCTCGAGCGCGCCGTATCCTCGCGGGTCTGGGCTTCAGCCGTGAGATGCAGGATCGACCCACCAAGAACTTCTCCGGTGGATGGAGAATGAGAGTTTCGCTCGCTAGAGCCCTGTACATAGAACCGACTCTGCTGTTACTAGACGAGCCTACGAACCATTTAGATCTGAACGCCGTGATCTGGTTAGACAATTACCTGCAAGGATGGAAGAAGACGTTACTAGTGGTATCTCACGATCAATCTTTCTTGGATAACGTTTGCAACGAAATCATACATTTAGATCAACAGAAATTGTTTTATTACAAAGGCAATTATTCGATGTTCAAGAAAATGTACGCACAGAAACGAAAGGAGATGATCAAGGAATATGAGAAACAAGAGAAGAGGATAAAGGAACTGAAGGCCCACGGACAGTCCAAGAAGCAAGCCGAAAAGAAGCAGAAGGATGCGTTGACGCGGAAACAGGAGAAGAACCGCAGCAAGGCGCAGCGCGAGGACGAGGACGGCGCCGCGCCCGTCGCCCTGCTGCAGCGGCCTAAGGAGTACATCGTCAAGTTCAGCTTCCCCGACCTGTCGCCGCTGCAGCCGCCCATCCTGGGACTGCACGACGTGGATTTCAACTTCCCCGGTCAGAAGCCTCTCTTCAAGGGCGTGAACTTCGGTATCGATCTTAATTCTCGTATCGCGATCGTGGGCCCCAACGGAGTCGGGAAATCGACGTTCCTGAAGCTACTCGTCGGCGAATTGACTCCTATACGCGGCGAACTGTTTAAGAACCACAGATTGAGGATAGGCAGATTCGACCAGCACTCTGGCGAGCACCTGACGGCGGAGGAGACGCCGGCGGAGTACCTGCAGCGCTTGTTCGGTCTGCAGTACGAGAAGGCGAGGAAGGCGCTCGGCACGTTCGGGCTGGCGAGCCACGCGCACATCATCAAGATGAAAGACCTCAGCGGCGGACAGAAGGCCCGCGTGGCGCTCGCCGAAATGACCCTCATGGGGCCCGACGTTATCATTCTGGATGAACCCACAAATAACTTGGACATCGAGAGTATAGACGCGTTGGCCGAAGCGATCAACGACTACAAGGGGGGCGTGGTCATAGTGTCCCACGACGAACGACTTATCAGAGAGACGGATTGCGCGCTCTACGTCATCGAAGATCAGACCATCAACGAGGTCGACGGTGACTTCGACGATTACCGTAAGGAGCTCCTCGAGAGCCTCGGGGAAACGGTCAACTCACCATCCATCATCGCTAACCAAGCAGTCCAACAATGA
Protein
MSKKKGNKKNQDLDDDFDEKPSVVNEKTELTSKNKGKGKKKNKTGDCSDSDEENIKKVPVSDEEDVPKPAAKKSQKKGKSKKKGADRSDDEEDDNKSIINNTTNVSSKPAKLAKKKKGKGKKDDDWSSEGSDVDLKMASDEDVAKPVGKKKTKGKKDDLPDEDSDKELKIEASEDDELVIPLVKSKGKHKKKNNMDDIEAELKKFEIKDPEHEEKSLKGKDKKKKRQEDESIVEEKDDDKVTELPSKEKEDSVEKKLSHKEKKKLKKQQEYEKQMELMTKKGGQGHSELDANFTVSQAQKTAGQMAALENAVDIKVENFSISAKGKDLFLNANLLIANGRRYGLVGPNGHGKTTLLRHLAQRAFPLPPHIDILLCEQEVTASDMSAVDTLLESDVKRTELLKECKELEAETEKGNLSKQDRLNEVYAELKAIGADAAEPRARRILAGLGFSREMQDRPTKNFSGGWRMRVSLARALYIEPTLLLLDEPTNHLDLNAVIWLDNYLQGWKKTLLVVSHDQSFLDNVCNEIIHLDQQKLFYYKGNYSMFKKMYAQKRKEMIKEYEKQEKRIKELKAHGQSKKQAEKKQKDALTRKQEKNRSKAQREDEDGAAPVALLQRPKEYIVKFSFPDLSPLQPPILGLHDVDFNFPGQKPLFKGVNFGIDLNSRIAIVGPNGVGKSTFLKLLVGELTPIRGELFKNHRLRIGRFDQHSGEHLTAEETPAEYLQRLFGLQYEKARKALGTFGLASHAHIIKMKDLSGGQKARVALAEMTLMGPDVIILDEPTNNLDIESIDALAEAINDYKGGVVIVSHDERLIRETDCALYVIEDQTINEVDGDFDDYRKELLESLGETVNSPSIIANQAVQQ
Summary
Uniprot
EMBL
Proteomes
PRIDE
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
PDB
5ZXD
E-value=0,
Score=1648
Ontologies
GO
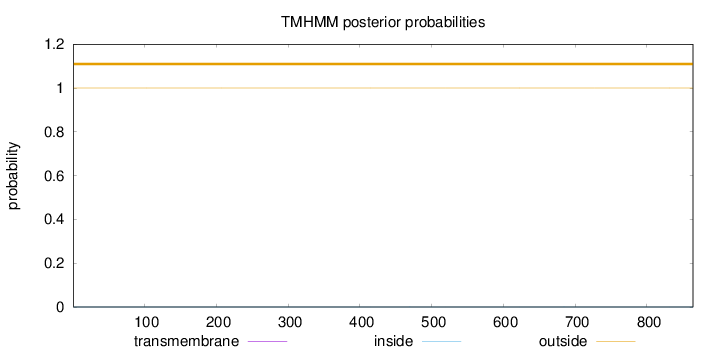
Topology
Length:
864
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00112
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00037
outside
1 - 864
Population Genetic Test Statistics
Pi
213.550416
Theta
155.73891
Tajima's D
1.384097
CLR
0.385062
CSRT
0.761461926903655
Interpretation
Uncertain
