Gene
KWMTBOMO08813
Pre Gene Modal
BGIBMGA007870
Annotation
PREDICTED:_probable_ATP-dependent_RNA_helicase_CG8611_[Bombyx_mori]
Full name
RNA helicase
Location in the cell
Mitochondrial Reliability : 1.828 Nuclear Reliability : 2.065
Sequence
CDS
ATGGATGATTTACAGTTAAACATATCTGTTTCACAAGGGAGTAAGAAGAAAGCAACATTGAAAAATAATGTATCAGAATATAAGTTTAAGTCTAAACCTACGTTTACTGGCAAAATTATAGCTAGAAAAAGGCCCGTAAACCTCGGGCAGAAGAATATAGATACTAAATCAGTTGATGATGGACCAAAGAAAAAGATTCTTAAAGCTTCACAAACACAAACAGGTGATTTAGCAACAAATTTAAGAGATCTTAGTGAAAACAAAGGAAAATTCCAAGGGAAGAACTATCAAGATCTCAGTGATGATATTCAAATGAAACAAAAGCCAAGAACAGGTGGATGGATATCTTCTCTTTTTAAAAATAATCCTGATGTTCCACGTATTGGGCAGCGTGCAGTCAAACCAATTGTTGAAAAAGTCTTTACTAGTAAAACATTTTCCGAGTTAGATATACATCCTCATAGTATTGCAAATTTACAACAGAATTTAGGTCTCAAAGAACTTATGACTGTACAACAAAATGCTATACCAGTTATTTTATGTGGACGAGATGTACTTATTAGGTCTCAGACAGGCTCAGGAAAAACCTTAGCATATGCTTTGCCAATAATAGAAGGACTTCAGGCGATAAGACCTAAAATAAATAGACATGACGGAATTCGGGCCATCATTGTTGTTCCGACACGTGAATTGGCAGTACAAACTTATGAATTGCTTGTTAAGCTTGTTAAGCCTTTTACCTGGATCGTACCTGGTCTTTTAAGTGGTGGTCAAAAACGGAAAGCAGAAAAAGCTAGGCTAAGAAAAGGATTAAGCATCCTTGTGGGAACACCGGGAAGGATAAATGACCACTTGAGGCATACAAAGTCTCTGAATTTTGATAAAACAGGTTGTTTAATTTTAGATGAAGCTGATAGGCTCTTGGATATGGGATATGAAAAAGACGTAGCTGCGATCGTCAAAGCTATTGAGGATCACAAAAAAGCTGCCACATACGATCCTATTGCTATGATTAAACAAAATGCAAAGAAGACGATCATAGATGATGATGAGTCTAAGCAAGTAGAACAGATTGATAATAAAGATGACTCGGTTGTTGCTAAACATCATTTGACTGAAGTGTTTCTTGGTAAAGACAGACAAACGATCTTATTATCTGCTACACTCACTAAGGCTGTAGAGAATTTAGCAGGAATCACAATGGTTGATCCTGTTTTCATTGATACATCCGAGGGAAAAGCAGTAGTTGCTGATTCTTTGAAACTACATACTAGCAATGTCATTAATAAAGAAGATGTATCAAAGGAAAAGAATGCAGCAATAGAGATCGAAACTGGAAATGAAATTAATATTAAGAAATCAAAAGAAAAAATATCCGTAAAGTCCGAAGAAGGCAATAAAACTGCTCTAGAAGCCAGAAGAGGTCTGAAATCCTTTGGCGGTATAAATAATTCTGAATTATTAAGTTCTAATACTAGTAAACTTGATAATGTAAATGAGAATAATATAACAAAAGATGTTAAAGACGAAACAAATAGTGATAGTGACTCTGACTATGAATATTTCAAAGTGCAAAAAGAACTTGAATCGAAAAAAGTAAGCGTGGAAACTATAAACGACGTAATAAAACCATCGGAAGTTGGCAACACTTTTGTTGAAGCTATAAAGACTGCAATAGTTGAAGATGAACTAGTGCTGCCCGCAACAGTGAATCAGACATTTTTAATTGTCCCAATGAAACTACGATTAGTCACCCTTTGTTCGTTGATCGTCGAACATTGTGTACTGAATAAGAAAGGCGGTAAAATGATTGTTTTTATGGCAACACTAGAGATGGTCGATTATCACTCCGAATTAATTGAGAATGTTTTAACTGGGAACAATGTAAAGGTAAAGAAATCTGGAAATGAAAAGAACAAAGCTAAGAAAAGAAAGGCCAACGATAATGAAAATCAGGATAAATCGGAATCGTCGGATTCGGAATTTGAAATTGATTATCAAGAGGCAAACGAAGGCGGCCTAGTGCCAGTCGACTTGGAGATGTTTAGTCTCCACGGCTCGATGCCGCACGAAAGGAGAATGGAAGTGTTCAAACAGTTTCGGACGGCCAGAAGCGGTGTTTTAATCTGTACGGATGTGGCGGCCCGTGGCTTGGACGTGCCTCGAGTGGACCTCGTCTTGCAGTACTGCGCACCCGCATCCGCAACTGATTATGTTCACAGGGTTGGGCGTACCGGACGAGCGGCGCGTATCGGTGCTGCGGTAATGTTTCTCTTACCGAACGAAGCGGATTTTGTTCGATATTTAGAACAAAAACGTATTAGGCTGCGGCAAGACGATGAATCGAAATGTCTGGAAGCGTTACGTAGCGTTGCACCGGCGGCGCCGAGCGTGCAACGCGCCGCCATCGCGCTACAAGCCCGCCTCGAACACACCGCGCACAGCTCAAAGGAATGGCTCGCTAGGGCCAGTCGAGCGTACACATCATGGGTGCGGTTTTACTCGGGCTACCCTCGCGACGTGCGTCAGTACTTGGATCCCAAACTGTTGCACCTCGGTCACGCGGCCAAAGCCTTCGCTTTGAGAGACACCCCTGCGGCCCTCGCCAGGAGAACTAAGTCCGATCCTAAACTGAAGAAAGAGAGACCCGTCAACAGACTTACAGTCCACGACGAAGAAGAAAGGAAGAGGCCCGGCTTCCCCAAAGTGAAGATCGGTTTTAATAGCAGAGTTATGAATAAGCAAAGTTCAATGCACACGGCCAGTGAATTCGATAGCGGACTGCCCCCACTTGAAAATTCATACAAAAAGAAAAAGAAAAACTGA
Protein
MDDLQLNISVSQGSKKKATLKNNVSEYKFKSKPTFTGKIIARKRPVNLGQKNIDTKSVDDGPKKKILKASQTQTGDLATNLRDLSENKGKFQGKNYQDLSDDIQMKQKPRTGGWISSLFKNNPDVPRIGQRAVKPIVEKVFTSKTFSELDIHPHSIANLQQNLGLKELMTVQQNAIPVILCGRDVLIRSQTGSGKTLAYALPIIEGLQAIRPKINRHDGIRAIIVVPTRELAVQTYELLVKLVKPFTWIVPGLLSGGQKRKAEKARLRKGLSILVGTPGRINDHLRHTKSLNFDKTGCLILDEADRLLDMGYEKDVAAIVKAIEDHKKAATYDPIAMIKQNAKKTIIDDDESKQVEQIDNKDDSVVAKHHLTEVFLGKDRQTILLSATLTKAVENLAGITMVDPVFIDTSEGKAVVADSLKLHTSNVINKEDVSKEKNAAIEIETGNEINIKKSKEKISVKSEEGNKTALEARRGLKSFGGINNSELLSSNTSKLDNVNENNITKDVKDETNSDSDSDYEYFKVQKELESKKVSVETINDVIKPSEVGNTFVEAIKTAIVEDELVLPATVNQTFLIVPMKLRLVTLCSLIVEHCVLNKKGGKMIVFMATLEMVDYHSELIENVLTGNNVKVKKSGNEKNKAKKRKANDNENQDKSESSDSEFEIDYQEANEGGLVPVDLEMFSLHGSMPHERRMEVFKQFRTARSGVLICTDVAARGLDVPRVDLVLQYCAPASATDYVHRVGRTGRAARIGAAVMFLLPNEADFVRYLEQKRIRLRQDDESKCLEALRSVAPAAPSVQRAAIALQARLEHTAHSSKEWLARASRAYTSWVRFYSGYPRDVRQYLDPKLLHLGHAAKAFALRDTPAALARRTKSDPKLKKERPVNRLTVHDEEERKRPGFPKVKIGFNSRVMNKQSSMHTASEFDSGLPPLENSYKKKKKN
Summary
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Similarity
Belongs to the DEAD box helicase family.
Uniprot
EC Number
3.6.4.13
EMBL
Proteomes
Interpro
SUPFAM
SSF52540
SSF52540
CDD
ProteinModelPortal
PDB
6EM5
E-value=1.49648e-29,
Score=327
Ontologies
GO
Topology
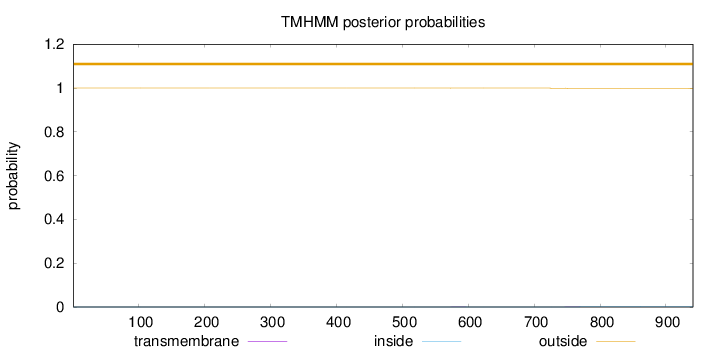
Length:
941
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0320500000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00009
outside
1 - 941
Population Genetic Test Statistics
Pi
271.294304
Theta
181.676843
Tajima's D
1.451293
CLR
0.52548
CSRT
0.772911354432278
Interpretation
Uncertain
