Gene
KWMTBOMO08809
Annotation
PREDICTED:_dynamin_isoform_X2_[Bombyx_mori]
Location in the cell
Cytoskeletal Reliability : 1.859 Nuclear Reliability : 1.38
Sequence
CDS
ATGGCTGGAAATTTTGGAATGCAGCAGTTGATTCCTATCGTAAACAAATTACAAGATGCATTTGTACAAATGGGCGTGCACATGTCCTTAGATCTTCCACAAATCGCTGTTGTGGGTGGTCAGTCGGCAGGAAAGAGTTCTGTTTTAGAGAATTTTGTCGGGAGGGATTTTTTACCAAGAGGCTCTGGCATTGTCACACGTCGTCCATTAATTCTGCAGCTTATAAATGGAAATACAGAATATGCAGAATTCTTACACTGCAAGGGAAAGAAGTTCACGGACTTCAATGAAGTTCGAGGAGAGATCGAGGCGGAGACAGATCGTGTTACTGGTTCCAACAAGGGTATATCTCCTGTACCTATTAACCTGCGTGTTTATTCCCCGAATGTGCTCAACCTGACGCTGATTGATTTGCCGGGTCTGACAAAAGTGCCAATCGGTGATCAGCCTATAGATATCGAGCAGCAAATCAAAGGGATGATATTCCAGTTCATCAGACGCGAATCATGCCTTATCCTGGCAGTTACTCCGGCCAACACGGATTTGGCCAACAGCGATGCTCTGAAGCTTGCCAAAGAAGTAGACCCACAAGGATTGCGTACTATCGGCGTAATAACAAAATTAGATTTGATGGACGAGGGTACCGATGCGCGGGACGTGCTCGAGAACAAGCTGCTGCCGCTGCGCCGAGGCTACATCGGAGTCGTCAACCGATCGCAGAAAGACATTGACGGTCGCAAGGACATAGCCGCTGCACTTGCGGCTGAGAGAAAATTCTTCCTTAGCCACCCGTCATACCGTCATTTGGCCGATCGTCTCGGTACCCCTTACCTACAGCGAGTGTTGAACCAACAGCTCACCAACCATATCCGCGACACTCTGCCCAGTCTCCGTGACAAGTTGCAGAAACAACTACTGACCTTGGAGAAGGACGTCGACCAGTACAAGCATTTCCGACCCGATGACCCGTCTATTAAAACTAAGGCCATGTTGCAAATGATTCAGACTTTGCAAACCGATTTTGAACGCACCATCGAAGGTTCCGGATCGGCGCAGATCAACACCAACGAATTGTCCGGTGGCGCTAAGATCAACAGGCTTTTCCACGAACGTTTTCCTTTTGAAATTGTCAAAATGGAGTTTGACGAAAAGGAACTGCGCCGCGAGATCGCGTTCGCCATTCGAAATATCCACGGTATCCGAGTGGGTTTGTTCACACCGGATATGGCGTTTGAGGCTATCGTGAAAAAGCAGATCGGACGTCTGAAGGAGCCGTGCCTGAAGTGCGTCGATCTGGTTGTGCAGGAGCTGTCCAACGTTGTACGGATTTGCACCGAACGAATGTCTCGTTACCCGCGCCTGCGCGAAGAGACGGAGCGCATCATAATGTCGCACGTGCGCTCGCGGGAGCAGATGTGCAAGGATCAGCTGGTGCTGCTCATCGACTGCGAACTGGCTTATATGAATACCAATCATGAAGATTTCATTGGATTCGCGAACGCACAAAATCAGTCTGAGAACTCTGCGAAGTCAGGACATCGAGCGCTCGGAAATCAGGTGATCCGAAAAGGATACATGTGCATTCACAATCTGGGAATTATGAAGGGAGGTTCCCGCGACTACTGGTTCGTACTGACTTCTGAGAGCATCTCGTGGTACAAGGATGAAGAGGAGCGGGAGAAGAAGTACATGTTGCCCCTCGACGGGCTCAAGTTGAGAGATCTCGAGCAGGGCTTCATGTCGCGGCGACACATGTTCGCACTCTTCAACCCTGAAGGCAGAAATGTTTACAAGGATTACAAGCAATTGGAGTTGTCGTGTGAAACTCAAGATGACGTCGATTCGTGGAAGGCTTCGTTCCTGCGCGCGGGAGTCTACCCCGAGAAGACATCGGAGGCGGCCAACGGCGACGAGAATTCGGACAGCACAGGCACCAGCAGTATGGACCCGCAGTTAGAGAGGCAGGTGGAGACCATCCGTAATCTGGTGGATTCGTACATGCGCATCGTGACGAAGACAACGCGAGATCTCGTCCCCAAGACAATCATGATGATGATTATCAATAATGCCAAAGACTTCATTAATGGAGAATTACTCGCGCATCTGTATGCGTCCGGGGATCAGTCTCAAATGATGGAAGAGTCTCCCGAGGAGGCACTGAAGCGTGAAGAGATGTTGCGCATGTACCACGCGTGCAAGGAGGCGCTGCACATCATCGGCGACGTGTCCATGGCCACCGTGAGCACGCCCGTCCCGCCGCCCGTCAAGAACGACTGGCTCGAGAGCAGACTCGACTCCAACCCGAGACTGTCGCCGCCCTCGCCCGGCGGCCCGCGCCGCGCCGCACCCGCGCAGCAAGGCACGTACACGCTCTGA
Protein
MAGNFGMQQLIPIVNKLQDAFVQMGVHMSLDLPQIAVVGGQSAGKSSVLENFVGRDFLPRGSGIVTRRPLILQLINGNTEYAEFLHCKGKKFTDFNEVRGEIEAETDRVTGSNKGISPVPINLRVYSPNVLNLTLIDLPGLTKVPIGDQPIDIEQQIKGMIFQFIRRESCLILAVTPANTDLANSDALKLAKEVDPQGLRTIGVITKLDLMDEGTDARDVLENKLLPLRRGYIGVVNRSQKDIDGRKDIAAALAAERKFFLSHPSYRHLADRLGTPYLQRVLNQQLTNHIRDTLPSLRDKLQKQLLTLEKDVDQYKHFRPDDPSIKTKAMLQMIQTLQTDFERTIEGSGSAQINTNELSGGAKINRLFHERFPFEIVKMEFDEKELRREIAFAIRNIHGIRVGLFTPDMAFEAIVKKQIGRLKEPCLKCVDLVVQELSNVVRICTERMSRYPRLREETERIIMSHVRSREQMCKDQLVLLIDCELAYMNTNHEDFIGFANAQNQSENSAKSGHRALGNQVIRKGYMCIHNLGIMKGGSRDYWFVLTSESISWYKDEEEREKKYMLPLDGLKLRDLEQGFMSRRHMFALFNPEGRNVYKDYKQLELSCETQDDVDSWKASFLRAGVYPEKTSEAANGDENSDSTGTSSMDPQLERQVETIRNLVDSYMRIVTKTTRDLVPKTIMMMIINNAKDFINGELLAHLYASGDQSQMMEESPEEALKREEMLRMYHACKEALHIIGDVSMATVSTPVPPPVKNDWLESRLDSNPRLSPPSPGGPRRAAPAQQGTYTL
Summary
Similarity
Belongs to the TRAFAC class dynamin-like GTPase superfamily. Dynamin/Fzo/YdjA family.
Uniprot
A0A2A4K370
A0A2A4K3U0
A0A2A4K466
A0A3S2P6X9
A0A194PRE1
A0A1E1W6Q7
+ More
D6X4J4 E2AA61 V9I9H2 A0A1W4WLA0 E2B692 A0A151ILI7 A0A195BYX1 A0A151IYR7 V9I7S1 A0A195FXL3 A0A0M8ZQM5 A0A151XGD9 A0A1Y1JZ95 A0A026WXD5 A0A1Y1K0B7 A0A1Y1JW64 A0A0P4VU42 A0A154PJG1 V9I9Y2 A0A088A6T0 A0A224XIG2 V9I7M3 A0A0C9PL23 A0A023F5T5 U4UBD3 A0A3T0D1N7 A0A069DX71 A0A158P3J0 N6TWX6 A0A232EYE3 K7IZQ3 A0A2A3EBL3 A0A1B6G972 A0A067RG47 A0A310SP26 A0A3L8DI49 A0A0A9X2I0 A0A146LZJ0 A0A1B6F7D2 A0A0A9WV52 A0A1B6GYH2 A0A1B6FZW1 A0A1B6M459 A0A1B6EZ89 A0A1B6LWT0 A0A0L7QPB4 E0VWF3 A0A1L8DW16 A0A1L8DVI7 A0A1B6L9E6 A0A1B6KTY4 T1I8U2 Q172Q6 Q172Q5 X1XTW3 A0A1Q3G459 A0A084VRC2 A0A2M3Z3A4 A0A182F7D9 A0A2M4A4P7 A0A182N637 A0A2M4BDG2 A0A0A1WPC2 A0A182Y4B0 A0A182QYT8 A0A1D2NFF0 A0A2M4A4L7 A0A2M4BDH6 A0A0A1WMG6 A0A182W067 A0A182P5T2 A0A0A1WHV0 U5EZL6 A0A182J339 A0A182RNQ9 A0A2M4CLP8 A0A0A1XF50 A0A182VAR5 A0A182JTF7 W8BD40 A0A0K8VD42 W8C039 W8B8L3 A0A034VHJ4 A0A182WRJ4 W8BD65 A0A182HU83 W8B1H6 Q5TU35 A0A034VLU2 A0A034VJV9 W5JEG4 A0A0V0GAN7 A0A182HU82
D6X4J4 E2AA61 V9I9H2 A0A1W4WLA0 E2B692 A0A151ILI7 A0A195BYX1 A0A151IYR7 V9I7S1 A0A195FXL3 A0A0M8ZQM5 A0A151XGD9 A0A1Y1JZ95 A0A026WXD5 A0A1Y1K0B7 A0A1Y1JW64 A0A0P4VU42 A0A154PJG1 V9I9Y2 A0A088A6T0 A0A224XIG2 V9I7M3 A0A0C9PL23 A0A023F5T5 U4UBD3 A0A3T0D1N7 A0A069DX71 A0A158P3J0 N6TWX6 A0A232EYE3 K7IZQ3 A0A2A3EBL3 A0A1B6G972 A0A067RG47 A0A310SP26 A0A3L8DI49 A0A0A9X2I0 A0A146LZJ0 A0A1B6F7D2 A0A0A9WV52 A0A1B6GYH2 A0A1B6FZW1 A0A1B6M459 A0A1B6EZ89 A0A1B6LWT0 A0A0L7QPB4 E0VWF3 A0A1L8DW16 A0A1L8DVI7 A0A1B6L9E6 A0A1B6KTY4 T1I8U2 Q172Q6 Q172Q5 X1XTW3 A0A1Q3G459 A0A084VRC2 A0A2M3Z3A4 A0A182F7D9 A0A2M4A4P7 A0A182N637 A0A2M4BDG2 A0A0A1WPC2 A0A182Y4B0 A0A182QYT8 A0A1D2NFF0 A0A2M4A4L7 A0A2M4BDH6 A0A0A1WMG6 A0A182W067 A0A182P5T2 A0A0A1WHV0 U5EZL6 A0A182J339 A0A182RNQ9 A0A2M4CLP8 A0A0A1XF50 A0A182VAR5 A0A182JTF7 W8BD40 A0A0K8VD42 W8C039 W8B8L3 A0A034VHJ4 A0A182WRJ4 W8BD65 A0A182HU83 W8B1H6 Q5TU35 A0A034VLU2 A0A034VJV9 W5JEG4 A0A0V0GAN7 A0A182HU82
Pubmed
EMBL
NWSH01000179
PCG78697.1
PCG78696.1
PCG78698.1
RSAL01000266
RVE43262.1
+ More
KQ459596 KPI95523.1 GDQN01008523 JAT82531.1 KQ971380 EEZ97258.2 GL438029 EFN69699.1 JR036359 AEY57106.1 GL445930 EFN88785.1 KQ977133 KYN05491.1 KQ976394 KYM93118.1 KQ980744 KYN13711.1 JR036358 AEY57105.1 KQ981161 KYN45410.1 KQ435899 KOX69135.1 KQ982174 KYQ59310.1 GEZM01099026 JAV53561.1 KK107088 EZA59804.1 GEZM01099027 JAV53560.1 GEZM01099028 JAV53559.1 GDKW01000433 JAI56162.1 KQ434936 KZC11967.1 JR036357 AEY57104.1 GFTR01008206 JAW08220.1 JR036356 AEY57103.1 GBYB01001703 JAG71470.1 GBBI01002168 JAC16544.1 KB632184 ERL89643.1 MH143574 AZT88971.1 GBGD01000483 JAC88406.1 ADTU01008108 ADTU01008109 ADTU01008110 ADTU01008111 ADTU01008112 APGK01048488 KB741112 ENN73805.1 NNAY01001646 OXU23329.1 KZ288312 PBC28441.1 GECZ01010795 JAS58974.1 KK852705 KDR18024.1 KQ760631 OAD59709.1 QOIP01000008 RLU19508.1 GBHO01032294 GBRD01010188 JAG11310.1 JAG55636.1 GDHC01006497 JAQ12132.1 GECZ01023715 JAS46054.1 GBHO01032299 GBHO01032293 GBRD01010190 JAG11305.1 JAG11311.1 JAG55634.1 GECZ01002294 JAS67475.1 GECZ01014059 JAS55710.1 GEBQ01009265 JAT30712.1 GECZ01026541 JAS43228.1 GEBQ01011860 JAT28117.1 KQ414819 KOC60478.1 DS235819 EEB17704.1 GFDF01003481 JAV10603.1 GFDF01003633 JAV10451.1 GEBQ01019823 JAT20154.1 GEBQ01025099 JAT14878.1 ACPB03003517 CH477432 EAT41041.1 EAT41042.1 ABLF02030849 ABLF02030855 ABLF02030856 GFDL01000457 JAV34588.1 ATLV01015604 KE525023 KFB40516.1 GGFM01002250 MBW23001.1 GGFK01002379 MBW35700.1 GGFJ01001902 MBW51043.1 GBXI01013590 JAD00702.1 AXCN02000193 LJIJ01000058 ODN03957.1 GGFK01002433 MBW35754.1 GGFJ01001954 MBW51095.1 GBXI01017229 GBXI01014240 JAC97062.1 JAD00052.1 GBXI01015855 JAC98436.1 GANO01000564 JAB59307.1 GGFL01002084 MBW66262.1 GBXI01005179 JAD09113.1 GAMC01011622 JAB94933.1 GDHF01015526 JAI36788.1 GAMC01011621 JAB94934.1 GAMC01011618 JAB94937.1 GAKP01016203 GAKP01016201 GAKP01016196 GAKP01016195 JAC42756.1 GAMC01011619 GAMC01011617 JAB94938.1 APCN01000612 GAMC01011620 GAMC01011616 JAB94935.1 AAAB01008859 EAL40783.4 GAKP01016202 GAKP01016199 GAKP01016194 GAKP01016193 JAC42750.1 GAKP01016204 GAKP01016200 GAKP01016198 GAKP01016197 JAC42754.1 ADMH02001371 ETN62747.1 GECL01001199 JAP04925.1
KQ459596 KPI95523.1 GDQN01008523 JAT82531.1 KQ971380 EEZ97258.2 GL438029 EFN69699.1 JR036359 AEY57106.1 GL445930 EFN88785.1 KQ977133 KYN05491.1 KQ976394 KYM93118.1 KQ980744 KYN13711.1 JR036358 AEY57105.1 KQ981161 KYN45410.1 KQ435899 KOX69135.1 KQ982174 KYQ59310.1 GEZM01099026 JAV53561.1 KK107088 EZA59804.1 GEZM01099027 JAV53560.1 GEZM01099028 JAV53559.1 GDKW01000433 JAI56162.1 KQ434936 KZC11967.1 JR036357 AEY57104.1 GFTR01008206 JAW08220.1 JR036356 AEY57103.1 GBYB01001703 JAG71470.1 GBBI01002168 JAC16544.1 KB632184 ERL89643.1 MH143574 AZT88971.1 GBGD01000483 JAC88406.1 ADTU01008108 ADTU01008109 ADTU01008110 ADTU01008111 ADTU01008112 APGK01048488 KB741112 ENN73805.1 NNAY01001646 OXU23329.1 KZ288312 PBC28441.1 GECZ01010795 JAS58974.1 KK852705 KDR18024.1 KQ760631 OAD59709.1 QOIP01000008 RLU19508.1 GBHO01032294 GBRD01010188 JAG11310.1 JAG55636.1 GDHC01006497 JAQ12132.1 GECZ01023715 JAS46054.1 GBHO01032299 GBHO01032293 GBRD01010190 JAG11305.1 JAG11311.1 JAG55634.1 GECZ01002294 JAS67475.1 GECZ01014059 JAS55710.1 GEBQ01009265 JAT30712.1 GECZ01026541 JAS43228.1 GEBQ01011860 JAT28117.1 KQ414819 KOC60478.1 DS235819 EEB17704.1 GFDF01003481 JAV10603.1 GFDF01003633 JAV10451.1 GEBQ01019823 JAT20154.1 GEBQ01025099 JAT14878.1 ACPB03003517 CH477432 EAT41041.1 EAT41042.1 ABLF02030849 ABLF02030855 ABLF02030856 GFDL01000457 JAV34588.1 ATLV01015604 KE525023 KFB40516.1 GGFM01002250 MBW23001.1 GGFK01002379 MBW35700.1 GGFJ01001902 MBW51043.1 GBXI01013590 JAD00702.1 AXCN02000193 LJIJ01000058 ODN03957.1 GGFK01002433 MBW35754.1 GGFJ01001954 MBW51095.1 GBXI01017229 GBXI01014240 JAC97062.1 JAD00052.1 GBXI01015855 JAC98436.1 GANO01000564 JAB59307.1 GGFL01002084 MBW66262.1 GBXI01005179 JAD09113.1 GAMC01011622 JAB94933.1 GDHF01015526 JAI36788.1 GAMC01011621 JAB94934.1 GAMC01011618 JAB94937.1 GAKP01016203 GAKP01016201 GAKP01016196 GAKP01016195 JAC42756.1 GAMC01011619 GAMC01011617 JAB94938.1 APCN01000612 GAMC01011620 GAMC01011616 JAB94935.1 AAAB01008859 EAL40783.4 GAKP01016202 GAKP01016199 GAKP01016194 GAKP01016193 JAC42750.1 GAKP01016204 GAKP01016200 GAKP01016198 GAKP01016197 JAC42754.1 ADMH02001371 ETN62747.1 GECL01001199 JAP04925.1
Proteomes
UP000218220
UP000283053
UP000053268
UP000007266
UP000000311
UP000192223
+ More
UP000008237 UP000078542 UP000078540 UP000078492 UP000078541 UP000053105 UP000075809 UP000053097 UP000076502 UP000005203 UP000030742 UP000005205 UP000019118 UP000215335 UP000002358 UP000242457 UP000027135 UP000279307 UP000053825 UP000009046 UP000015103 UP000008820 UP000007819 UP000030765 UP000069272 UP000075884 UP000076408 UP000075886 UP000094527 UP000075920 UP000075885 UP000075880 UP000075900 UP000075903 UP000075881 UP000076407 UP000075840 UP000007062 UP000000673
UP000008237 UP000078542 UP000078540 UP000078492 UP000078541 UP000053105 UP000075809 UP000053097 UP000076502 UP000005203 UP000030742 UP000005205 UP000019118 UP000215335 UP000002358 UP000242457 UP000027135 UP000279307 UP000053825 UP000009046 UP000015103 UP000008820 UP000007819 UP000030765 UP000069272 UP000075884 UP000076408 UP000075886 UP000094527 UP000075920 UP000075885 UP000075880 UP000075900 UP000075903 UP000075881 UP000076407 UP000075840 UP000007062 UP000000673
Interpro
IPR003130
GED
+ More
IPR027417 P-loop_NTPase
IPR001849 PH_domain
IPR030381 G_DYNAMIN_dom
IPR011993 PH-like_dom_sf
IPR019762 Dynamin_GTPase_CS
IPR001401 Dynamin_GTPase
IPR027741 DNM1
IPR000375 Dynamin_central
IPR020850 GED_dom
IPR022812 Dynamin_SF
IPR004148 BAR_dom
IPR036028 SH3-like_dom_sf
IPR027267 AH/BAR_dom_sf
IPR001452 SH3_domain
IPR027417 P-loop_NTPase
IPR001849 PH_domain
IPR030381 G_DYNAMIN_dom
IPR011993 PH-like_dom_sf
IPR019762 Dynamin_GTPase_CS
IPR001401 Dynamin_GTPase
IPR027741 DNM1
IPR000375 Dynamin_central
IPR020850 GED_dom
IPR022812 Dynamin_SF
IPR004148 BAR_dom
IPR036028 SH3-like_dom_sf
IPR027267 AH/BAR_dom_sf
IPR001452 SH3_domain
Gene 3D
CDD
ProteinModelPortal
A0A2A4K370
A0A2A4K3U0
A0A2A4K466
A0A3S2P6X9
A0A194PRE1
A0A1E1W6Q7
+ More
D6X4J4 E2AA61 V9I9H2 A0A1W4WLA0 E2B692 A0A151ILI7 A0A195BYX1 A0A151IYR7 V9I7S1 A0A195FXL3 A0A0M8ZQM5 A0A151XGD9 A0A1Y1JZ95 A0A026WXD5 A0A1Y1K0B7 A0A1Y1JW64 A0A0P4VU42 A0A154PJG1 V9I9Y2 A0A088A6T0 A0A224XIG2 V9I7M3 A0A0C9PL23 A0A023F5T5 U4UBD3 A0A3T0D1N7 A0A069DX71 A0A158P3J0 N6TWX6 A0A232EYE3 K7IZQ3 A0A2A3EBL3 A0A1B6G972 A0A067RG47 A0A310SP26 A0A3L8DI49 A0A0A9X2I0 A0A146LZJ0 A0A1B6F7D2 A0A0A9WV52 A0A1B6GYH2 A0A1B6FZW1 A0A1B6M459 A0A1B6EZ89 A0A1B6LWT0 A0A0L7QPB4 E0VWF3 A0A1L8DW16 A0A1L8DVI7 A0A1B6L9E6 A0A1B6KTY4 T1I8U2 Q172Q6 Q172Q5 X1XTW3 A0A1Q3G459 A0A084VRC2 A0A2M3Z3A4 A0A182F7D9 A0A2M4A4P7 A0A182N637 A0A2M4BDG2 A0A0A1WPC2 A0A182Y4B0 A0A182QYT8 A0A1D2NFF0 A0A2M4A4L7 A0A2M4BDH6 A0A0A1WMG6 A0A182W067 A0A182P5T2 A0A0A1WHV0 U5EZL6 A0A182J339 A0A182RNQ9 A0A2M4CLP8 A0A0A1XF50 A0A182VAR5 A0A182JTF7 W8BD40 A0A0K8VD42 W8C039 W8B8L3 A0A034VHJ4 A0A182WRJ4 W8BD65 A0A182HU83 W8B1H6 Q5TU35 A0A034VLU2 A0A034VJV9 W5JEG4 A0A0V0GAN7 A0A182HU82
D6X4J4 E2AA61 V9I9H2 A0A1W4WLA0 E2B692 A0A151ILI7 A0A195BYX1 A0A151IYR7 V9I7S1 A0A195FXL3 A0A0M8ZQM5 A0A151XGD9 A0A1Y1JZ95 A0A026WXD5 A0A1Y1K0B7 A0A1Y1JW64 A0A0P4VU42 A0A154PJG1 V9I9Y2 A0A088A6T0 A0A224XIG2 V9I7M3 A0A0C9PL23 A0A023F5T5 U4UBD3 A0A3T0D1N7 A0A069DX71 A0A158P3J0 N6TWX6 A0A232EYE3 K7IZQ3 A0A2A3EBL3 A0A1B6G972 A0A067RG47 A0A310SP26 A0A3L8DI49 A0A0A9X2I0 A0A146LZJ0 A0A1B6F7D2 A0A0A9WV52 A0A1B6GYH2 A0A1B6FZW1 A0A1B6M459 A0A1B6EZ89 A0A1B6LWT0 A0A0L7QPB4 E0VWF3 A0A1L8DW16 A0A1L8DVI7 A0A1B6L9E6 A0A1B6KTY4 T1I8U2 Q172Q6 Q172Q5 X1XTW3 A0A1Q3G459 A0A084VRC2 A0A2M3Z3A4 A0A182F7D9 A0A2M4A4P7 A0A182N637 A0A2M4BDG2 A0A0A1WPC2 A0A182Y4B0 A0A182QYT8 A0A1D2NFF0 A0A2M4A4L7 A0A2M4BDH6 A0A0A1WMG6 A0A182W067 A0A182P5T2 A0A0A1WHV0 U5EZL6 A0A182J339 A0A182RNQ9 A0A2M4CLP8 A0A0A1XF50 A0A182VAR5 A0A182JTF7 W8BD40 A0A0K8VD42 W8C039 W8B8L3 A0A034VHJ4 A0A182WRJ4 W8BD65 A0A182HU83 W8B1H6 Q5TU35 A0A034VLU2 A0A034VJV9 W5JEG4 A0A0V0GAN7 A0A182HU82
PDB
4UUK
E-value=0,
Score=3034
Ontologies
GO
GO:0072583
GO:0005525
GO:0003924
GO:0044327
GO:0016185
GO:0031410
GO:0005737
GO:0043197
GO:0098884
GO:0031623
GO:0050803
GO:0030424
GO:0008017
GO:0014069
GO:0045211
GO:0005886
GO:0098793
GO:0098844
GO:0045202
GO:0016020
GO:0005840
GO:0006412
GO:0008097
GO:0016817
GO:0008270
GO:0043565
GO:0016876
GO:0043039
PANTHER
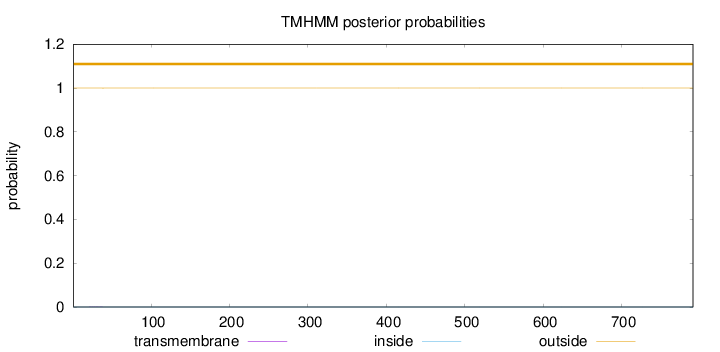
Topology
Length:
791
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00450999999999999
Exp number, first 60 AAs:
0.00302
Total prob of N-in:
0.00019
outside
1 - 791
Population Genetic Test Statistics
Pi
215.484773
Theta
174.432435
Tajima's D
0.623765
CLR
0.01747
CSRT
0.547222638868057
Interpretation
Uncertain
