Gene
KWMTBOMO08807
Pre Gene Modal
BGIBMGA007876
Annotation
PREDICTED:_CTL-like_protein_2_isoform_X2_[Bombyx_mori]
Full name
CTL-like protein 2
Location in the cell
PlasmaMembrane Reliability : 4.96
Sequence
CDS
ATGTACATTGGTTACTATGGTTATCAAAATGGTAACATTGAAAAACTTTTAGCTCCAATGGACTCGAACGGTAAGCGATGCGGCGTTGATTCTGGTGTTATAAATAGAAAATATTTATTATTTTTCAACTTAGCGAATTGTCTAAGGCCCGAGACACCGATTACTGGATGCCCTACTCCACAAGTTTGCGTAGAGGAATGTCCCCAAAGGACAGTAAAATTTATTGAACAGCTAACTTCGGCAAATTTCAATGAAATGAAACGAGAAATGGTGTGCATCGACGGCGTTAATACGGAAACAATGACATATAGCGACGCTTTGCAATACATGAACGAAGATAAATGTGCTGGCCTAATTTTACAAAGCCAGTCAGTGTTGTTTCGCTGCATCGGAGATCTATCCACCATCCCATGCAAGGGCCCACCCAAACCGACCACAAATTGGATGCAAACCGACGGAGACACTACATGTGTAAGAAATCCTAAGGAAGGACAGATGTCGTTGTTCAATCGTGCAACGGCTCTCGACACTTATATTGGCAGGATTGTCGCTAGCTTGGTGACCGCCTTCACGAAAAACGAAAGGGACACGCATATTCTATCATCAAAAATCGTCCAAGATCTGGTTCAATCTCGGTGGTACCTCGCTGGAGCTTTATTGGCAGTTGTCATTATCTGTTTCATTTATATAGTTTTACTGCGGTTTGTCATTACACCTATCGTCTGGGCTTCCATCGGAGGGCTCTTAGCGCTACTTATTTTTACGTCATACTTGTGCTATGGAAACTACGTATATTACCGTGATAATCCCATTCAACTTCACCAGACGACAAATCTCAAAGGATATGCGCAGTCTATATTCAGCAAGGCGTCGACGTGGATGGTCATATTAATTGTGACGGCCTTTCTAAGTTTCATACTTTTAGTAATGGTTGTATTTTTACGAAACCGCATCAGGATCGCCATTGCACTCATACGAGAGGGCAGCAGAGCATTGTCGGCGGTCAAATCAACAGTTTTATTGCCTATATTCCCATGGATATTTCAATGTGCTGTCATCGCCTATTGGCTGTTCTTGTTTATGTCCTTGCTGTCGATAGGAACGCCGATATTCAAGACTGCCAACTTAAAATCGGATACCAATTGCAAATGTGCAATTTATACCGAAGATAATATGGATTGTGATCCAAATAAATTTACACTCTCAAGTCGCGATTTAAGTGGCGGCCCTTGCCGACAGGCGACTTGCACCTTCCAAAGCTTTGAAACACCTCAGATCATCTTATATTTACATTTGGCCAACTTTCTCGGATTTTTTTGGACGATGTTCTTCATTACCGGTATTACCGACATGATACTCGCGAGCACCTTCGCCACTTGGTACTGGACTTTCCACAAAAGCGATCTTCCTTTCTTCACCCTCACGAGAGGCGCGTACAGAACCATAAGATACCACACGGGAACCGTGGCATTTGGCTCATTAATTATTGCCATTGTTCGAGTGATAAGAGTAATACTCGAGTACATTGATCACAAATTGAAAAAGTTCGAAAATCCCTTTACGAGATTCATAATGTGCTGTTGCAAATGTTTGTTCTGGTGCCTGGAGAGCTTCCTCAAGTTCATCAATAAGAACGCTTTTATAATGTGTGCGGTGCACGGACACAACTTCTGTAAAAGTGCTAGAGATGCGTTTTCCTTACTCCTAAGGAATGTTGTTAGAGTCGTTGTTTTAGACAAGGTGACCGATTTCATATTCTTCGTATCGAAGCTTCTGATATCAATAGGGATAGGCTTCGCAGTATACTATATTTTAGAATGGAATTTTACTTACGAAATCACGAAAGGCCAACGTTTAAATTACAACTACGTACCAGCCATAATACTGAGTGTCGCGACTTACATCATATGCACTGTGTTCTTCAATGTTTACTCTATGGCTGTTGATACTTTGTTCCTTTGTTTCCTTGAAGACTGCGAGAGAAATGATGGATCCCCCGAAAAACCATATTTTATGTCAAAAAATCTTATGAAAATACTTGGAAAAAGAAACAAACAGCTATAA
Protein
MYIGYYGYQNGNIEKLLAPMDSNGKRCGVDSGVINRKYLLFFNLANCLRPETPITGCPTPQVCVEECPQRTVKFIEQLTSANFNEMKREMVCIDGVNTETMTYSDALQYMNEDKCAGLILQSQSVLFRCIGDLSTIPCKGPPKPTTNWMQTDGDTTCVRNPKEGQMSLFNRATALDTYIGRIVASLVTAFTKNERDTHILSSKIVQDLVQSRWYLAGALLAVVIICFIYIVLLRFVITPIVWASIGGLLALLIFTSYLCYGNYVYYRDNPIQLHQTTNLKGYAQSIFSKASTWMVILIVTAFLSFILLVMVVFLRNRIRIAIALIREGSRALSAVKSTVLLPIFPWIFQCAVIAYWLFLFMSLLSIGTPIFKTANLKSDTNCKCAIYTEDNMDCDPNKFTLSSRDLSGGPCRQATCTFQSFETPQIILYLHLANFLGFFWTMFFITGITDMILASTFATWYWTFHKSDLPFFTLTRGAYRTIRYHTGTVAFGSLIIAIVRVIRVILEYIDHKLKKFENPFTRFIMCCCKCLFWCLESFLKFINKNAFIMCAVHGHNFCKSARDAFSLLLRNVVRVVVLDKVTDFIFFVSKLLISIGIGFAVYYILEWNFTYEITKGQRLNYNYVPAIILSVATYIICTVFFNVYSMAVDTLFLCFLEDCERNDGSPEKPYFMSKNLMKILGKRNKQL
Summary
Similarity
Belongs to the CTL (choline transporter-like) family.
Keywords
Complete proteome
Glycoprotein
Membrane
Reference proteome
Transmembrane
Transmembrane helix
Feature
chain CTL-like protein 2
Uniprot
H9JEC9
A0A194QME2
A0A3S2LS77
A0A2A4K470
A0A1E1W227
A0A212F0V1
+ More
A0A2A4K362 A0A2A4K4L2 A0A2A4K4W0 A0A194PRZ1 A0A1L8DEX3 A0A2H1WUM1 A0A1Y1JUX5 A0A1Q3FY28 A0A1Q3FXG9 A0A1Q3FY33 A0A1Q3FY17 A0A1L8DEM6 A0A1L8DEG2 A0A182NQ24 A0A023EV43 A0A0A1WF97 A0A2M4A2S3 A0A1I8Q561 D6WC46 A0A2M4CZP1 A0A1S4H3Q3 A0A139WME4 A0A139WM68 T1PF59 A0A1I8NC65 A0A1Q3G3N9 A0A1Q3G3L7 A0A1Q3G3L9 A0A182JP45 A0A182R922 A0A1Q3G3U6 A0A1Y1JW24 B4K750 A0A182YE22 A0A034W7A9 B4NBG0 A0A0M4EEH7 Q7PRJ0 Q17IP0 A0A182PBV3 A0A182JCJ5 A0A3B0JE03 W5JPK8 B3MT44 A0A2M4CQB0 B4M0B5 A0A1S4H4N4 B4JRR0 A0A2M3YYN7 B4G2H7 Q29AB8 A0A0P4VUD9 B4QZG9 C4XVJ5 Q9VAP3 A0A182W864 A0A224XKU8 A0A0K8W159 A0A0A9XR06 A0A146L112 A0A0A9XHQ7 B4PQ70 A0A0A9ZCJ5 A0A1B6JVG8 A0A0A9Z508 A0A1W4UIU2 B3P5L1 W8B7F8 A0A2M4A3G6 A0A1L8DDR7 A0A336LLH3 A0A1L8DDT3 A0A067R4F2 A0A1B6FU38 A0A0T6B8V2 A0A1B6D8K7 T1HK07 A0A1B6ECV3 A0A232F0V2 A0A2M4A3J7 K7IXB7 A0A0P5D418 A0A0P5DB75 A0A0N8DVF3 A0A0P5YYJ2 A0A0P5A1H5 A0A182X4Y8 E9FWB8 A0A2H8TYW2 J9K3H1 A0A2J7RKG8 A0A087UQP2 A0A0P6IHC9 A0A0P6GFU0
A0A2A4K362 A0A2A4K4L2 A0A2A4K4W0 A0A194PRZ1 A0A1L8DEX3 A0A2H1WUM1 A0A1Y1JUX5 A0A1Q3FY28 A0A1Q3FXG9 A0A1Q3FY33 A0A1Q3FY17 A0A1L8DEM6 A0A1L8DEG2 A0A182NQ24 A0A023EV43 A0A0A1WF97 A0A2M4A2S3 A0A1I8Q561 D6WC46 A0A2M4CZP1 A0A1S4H3Q3 A0A139WME4 A0A139WM68 T1PF59 A0A1I8NC65 A0A1Q3G3N9 A0A1Q3G3L7 A0A1Q3G3L9 A0A182JP45 A0A182R922 A0A1Q3G3U6 A0A1Y1JW24 B4K750 A0A182YE22 A0A034W7A9 B4NBG0 A0A0M4EEH7 Q7PRJ0 Q17IP0 A0A182PBV3 A0A182JCJ5 A0A3B0JE03 W5JPK8 B3MT44 A0A2M4CQB0 B4M0B5 A0A1S4H4N4 B4JRR0 A0A2M3YYN7 B4G2H7 Q29AB8 A0A0P4VUD9 B4QZG9 C4XVJ5 Q9VAP3 A0A182W864 A0A224XKU8 A0A0K8W159 A0A0A9XR06 A0A146L112 A0A0A9XHQ7 B4PQ70 A0A0A9ZCJ5 A0A1B6JVG8 A0A0A9Z508 A0A1W4UIU2 B3P5L1 W8B7F8 A0A2M4A3G6 A0A1L8DDR7 A0A336LLH3 A0A1L8DDT3 A0A067R4F2 A0A1B6FU38 A0A0T6B8V2 A0A1B6D8K7 T1HK07 A0A1B6ECV3 A0A232F0V2 A0A2M4A3J7 K7IXB7 A0A0P5D418 A0A0P5DB75 A0A0N8DVF3 A0A0P5YYJ2 A0A0P5A1H5 A0A182X4Y8 E9FWB8 A0A2H8TYW2 J9K3H1 A0A2J7RKG8 A0A087UQP2 A0A0P6IHC9 A0A0P6GFU0
Pubmed
EMBL
BABH01002889
KQ461195
KPJ06708.1
RSAL01000266
RVE43260.1
NWSH01000179
+ More
PCG78704.1 GDQN01010070 GDQN01005024 JAT80984.1 JAT86030.1 AGBW02011012 OWR47375.1 PCG78701.1 PCG78703.1 PCG78702.1 KQ459596 KPI95519.1 GFDF01009157 JAV04927.1 ODYU01011134 SOQ56682.1 GEZM01103029 GEZM01103028 GEZM01103027 GEZM01103026 GEZM01103025 GEZM01103023 JAV51600.1 GFDL01002578 JAV32467.1 GFDL01002758 JAV32287.1 GFDL01002577 JAV32468.1 GFDL01002588 JAV32457.1 GFDF01009249 JAV04835.1 GFDF01009248 JAV04836.1 GAPW01000278 JAC13320.1 GBXI01016613 GBXI01014997 JAC97678.1 JAC99294.1 GGFK01001772 MBW35093.1 KQ971316 EEZ98759.2 GGFL01006571 MBW70749.1 AAAB01008849 KYB28985.1 KYB28984.1 KA647324 AFP61953.1 GFDL01000631 JAV34414.1 GFDL01000647 JAV34398.1 GFDL01000642 JAV34403.1 GFDL01000609 JAV34436.1 GEZM01103024 JAV51605.1 CH933806 EDW16363.1 KRG02065.1 KRG02066.1 GAKP01008359 GAKP01008358 GAKP01008357 JAC50594.1 CH964232 EDW81124.2 CP012526 ALC46404.1 CH477238 EAT46520.1 OUUW01000005 SPP80594.1 ADMH02000477 ETN66327.1 CH902623 EDV30434.1 KPU72849.1 GGFL01003329 MBW67507.1 CH940650 EDW67277.1 CH916373 EDV94450.1 GGFM01000587 MBW21338.1 CH479179 EDW24022.1 CM000070 EAL27432.3 KRT00021.1 KRT00022.1 GDKW01000296 JAI56299.1 CM000364 EDX14807.1 BT088799 ACS16657.1 AE014297 GFTR01007777 JAW08649.1 GDHF01030697 GDHF01023730 GDHF01021624 GDHF01014652 GDHF01007493 GDHF01002436 JAI21617.1 JAI28584.1 JAI30690.1 JAI37662.1 JAI44821.1 JAI49878.1 GBHO01024069 JAG19535.1 GDHC01016571 JAQ02058.1 GBHO01024075 GBRD01010862 GDHC01004258 JAG19529.1 JAG54962.1 JAQ14371.1 CM000160 EDW98339.1 KRK04229.1 GBHO01004069 JAG39535.1 GECU01004508 JAT03199.1 GBHO01004070 GBRD01010861 JAG39534.1 JAG54963.1 CH954182 EDV53261.1 GAMC01013592 GAMC01013591 GAMC01013590 JAB92963.1 GGFK01001970 MBW35291.1 GFDF01009471 JAV04613.1 UFQS01000012 UFQT01000012 SSW97215.1 SSX17601.1 GFDF01009472 JAV04612.1 KK852703 KDR18085.1 GECZ01016075 JAS53694.1 LJIG01009097 KRT83761.1 GEDC01015308 JAS21990.1 ACPB03013231 ACPB03013232 ACPB03013233 ACPB03013234 GEDC01001594 JAS35704.1 NNAY01001328 OXU24345.1 GGFK01002046 MBW35367.1 AAZX01000026 GDIP01165727 JAJ57675.1 GDIP01158490 GDIP01145839 GDIP01064997 JAJ64912.1 GDIQ01088044 JAN06693.1 GDIP01053310 JAM50405.1 GDIP01217925 JAJ05477.1 GL732526 EFX88458.1 GFXV01007690 MBW19495.1 ABLF02039129 NEVH01002737 PNF41327.1 KK121074 KFM79681.1 GDIQ01011066 JAN83671.1 GDIQ01053817 JAN40920.1
PCG78704.1 GDQN01010070 GDQN01005024 JAT80984.1 JAT86030.1 AGBW02011012 OWR47375.1 PCG78701.1 PCG78703.1 PCG78702.1 KQ459596 KPI95519.1 GFDF01009157 JAV04927.1 ODYU01011134 SOQ56682.1 GEZM01103029 GEZM01103028 GEZM01103027 GEZM01103026 GEZM01103025 GEZM01103023 JAV51600.1 GFDL01002578 JAV32467.1 GFDL01002758 JAV32287.1 GFDL01002577 JAV32468.1 GFDL01002588 JAV32457.1 GFDF01009249 JAV04835.1 GFDF01009248 JAV04836.1 GAPW01000278 JAC13320.1 GBXI01016613 GBXI01014997 JAC97678.1 JAC99294.1 GGFK01001772 MBW35093.1 KQ971316 EEZ98759.2 GGFL01006571 MBW70749.1 AAAB01008849 KYB28985.1 KYB28984.1 KA647324 AFP61953.1 GFDL01000631 JAV34414.1 GFDL01000647 JAV34398.1 GFDL01000642 JAV34403.1 GFDL01000609 JAV34436.1 GEZM01103024 JAV51605.1 CH933806 EDW16363.1 KRG02065.1 KRG02066.1 GAKP01008359 GAKP01008358 GAKP01008357 JAC50594.1 CH964232 EDW81124.2 CP012526 ALC46404.1 CH477238 EAT46520.1 OUUW01000005 SPP80594.1 ADMH02000477 ETN66327.1 CH902623 EDV30434.1 KPU72849.1 GGFL01003329 MBW67507.1 CH940650 EDW67277.1 CH916373 EDV94450.1 GGFM01000587 MBW21338.1 CH479179 EDW24022.1 CM000070 EAL27432.3 KRT00021.1 KRT00022.1 GDKW01000296 JAI56299.1 CM000364 EDX14807.1 BT088799 ACS16657.1 AE014297 GFTR01007777 JAW08649.1 GDHF01030697 GDHF01023730 GDHF01021624 GDHF01014652 GDHF01007493 GDHF01002436 JAI21617.1 JAI28584.1 JAI30690.1 JAI37662.1 JAI44821.1 JAI49878.1 GBHO01024069 JAG19535.1 GDHC01016571 JAQ02058.1 GBHO01024075 GBRD01010862 GDHC01004258 JAG19529.1 JAG54962.1 JAQ14371.1 CM000160 EDW98339.1 KRK04229.1 GBHO01004069 JAG39535.1 GECU01004508 JAT03199.1 GBHO01004070 GBRD01010861 JAG39534.1 JAG54963.1 CH954182 EDV53261.1 GAMC01013592 GAMC01013591 GAMC01013590 JAB92963.1 GGFK01001970 MBW35291.1 GFDF01009471 JAV04613.1 UFQS01000012 UFQT01000012 SSW97215.1 SSX17601.1 GFDF01009472 JAV04612.1 KK852703 KDR18085.1 GECZ01016075 JAS53694.1 LJIG01009097 KRT83761.1 GEDC01015308 JAS21990.1 ACPB03013231 ACPB03013232 ACPB03013233 ACPB03013234 GEDC01001594 JAS35704.1 NNAY01001328 OXU24345.1 GGFK01002046 MBW35367.1 AAZX01000026 GDIP01165727 JAJ57675.1 GDIP01158490 GDIP01145839 GDIP01064997 JAJ64912.1 GDIQ01088044 JAN06693.1 GDIP01053310 JAM50405.1 GDIP01217925 JAJ05477.1 GL732526 EFX88458.1 GFXV01007690 MBW19495.1 ABLF02039129 NEVH01002737 PNF41327.1 KK121074 KFM79681.1 GDIQ01011066 JAN83671.1 GDIQ01053817 JAN40920.1
Proteomes
UP000005204
UP000053240
UP000283053
UP000218220
UP000007151
UP000053268
+ More
UP000075884 UP000095300 UP000007266 UP000095301 UP000075881 UP000075900 UP000009192 UP000076408 UP000007798 UP000092553 UP000007062 UP000008820 UP000075885 UP000075880 UP000268350 UP000000673 UP000007801 UP000008792 UP000001070 UP000008744 UP000001819 UP000000304 UP000000803 UP000075920 UP000002282 UP000192221 UP000008711 UP000027135 UP000015103 UP000215335 UP000002358 UP000076407 UP000000305 UP000007819 UP000235965 UP000054359
UP000075884 UP000095300 UP000007266 UP000095301 UP000075881 UP000075900 UP000009192 UP000076408 UP000007798 UP000092553 UP000007062 UP000008820 UP000075885 UP000075880 UP000268350 UP000000673 UP000007801 UP000008792 UP000001070 UP000008744 UP000001819 UP000000304 UP000000803 UP000075920 UP000002282 UP000192221 UP000008711 UP000027135 UP000015103 UP000215335 UP000002358 UP000076407 UP000000305 UP000007819 UP000235965 UP000054359
Interpro
SUPFAM
SSF52091
SSF52091
Gene 3D
ProteinModelPortal
H9JEC9
A0A194QME2
A0A3S2LS77
A0A2A4K470
A0A1E1W227
A0A212F0V1
+ More
A0A2A4K362 A0A2A4K4L2 A0A2A4K4W0 A0A194PRZ1 A0A1L8DEX3 A0A2H1WUM1 A0A1Y1JUX5 A0A1Q3FY28 A0A1Q3FXG9 A0A1Q3FY33 A0A1Q3FY17 A0A1L8DEM6 A0A1L8DEG2 A0A182NQ24 A0A023EV43 A0A0A1WF97 A0A2M4A2S3 A0A1I8Q561 D6WC46 A0A2M4CZP1 A0A1S4H3Q3 A0A139WME4 A0A139WM68 T1PF59 A0A1I8NC65 A0A1Q3G3N9 A0A1Q3G3L7 A0A1Q3G3L9 A0A182JP45 A0A182R922 A0A1Q3G3U6 A0A1Y1JW24 B4K750 A0A182YE22 A0A034W7A9 B4NBG0 A0A0M4EEH7 Q7PRJ0 Q17IP0 A0A182PBV3 A0A182JCJ5 A0A3B0JE03 W5JPK8 B3MT44 A0A2M4CQB0 B4M0B5 A0A1S4H4N4 B4JRR0 A0A2M3YYN7 B4G2H7 Q29AB8 A0A0P4VUD9 B4QZG9 C4XVJ5 Q9VAP3 A0A182W864 A0A224XKU8 A0A0K8W159 A0A0A9XR06 A0A146L112 A0A0A9XHQ7 B4PQ70 A0A0A9ZCJ5 A0A1B6JVG8 A0A0A9Z508 A0A1W4UIU2 B3P5L1 W8B7F8 A0A2M4A3G6 A0A1L8DDR7 A0A336LLH3 A0A1L8DDT3 A0A067R4F2 A0A1B6FU38 A0A0T6B8V2 A0A1B6D8K7 T1HK07 A0A1B6ECV3 A0A232F0V2 A0A2M4A3J7 K7IXB7 A0A0P5D418 A0A0P5DB75 A0A0N8DVF3 A0A0P5YYJ2 A0A0P5A1H5 A0A182X4Y8 E9FWB8 A0A2H8TYW2 J9K3H1 A0A2J7RKG8 A0A087UQP2 A0A0P6IHC9 A0A0P6GFU0
A0A2A4K362 A0A2A4K4L2 A0A2A4K4W0 A0A194PRZ1 A0A1L8DEX3 A0A2H1WUM1 A0A1Y1JUX5 A0A1Q3FY28 A0A1Q3FXG9 A0A1Q3FY33 A0A1Q3FY17 A0A1L8DEM6 A0A1L8DEG2 A0A182NQ24 A0A023EV43 A0A0A1WF97 A0A2M4A2S3 A0A1I8Q561 D6WC46 A0A2M4CZP1 A0A1S4H3Q3 A0A139WME4 A0A139WM68 T1PF59 A0A1I8NC65 A0A1Q3G3N9 A0A1Q3G3L7 A0A1Q3G3L9 A0A182JP45 A0A182R922 A0A1Q3G3U6 A0A1Y1JW24 B4K750 A0A182YE22 A0A034W7A9 B4NBG0 A0A0M4EEH7 Q7PRJ0 Q17IP0 A0A182PBV3 A0A182JCJ5 A0A3B0JE03 W5JPK8 B3MT44 A0A2M4CQB0 B4M0B5 A0A1S4H4N4 B4JRR0 A0A2M3YYN7 B4G2H7 Q29AB8 A0A0P4VUD9 B4QZG9 C4XVJ5 Q9VAP3 A0A182W864 A0A224XKU8 A0A0K8W159 A0A0A9XR06 A0A146L112 A0A0A9XHQ7 B4PQ70 A0A0A9ZCJ5 A0A1B6JVG8 A0A0A9Z508 A0A1W4UIU2 B3P5L1 W8B7F8 A0A2M4A3G6 A0A1L8DDR7 A0A336LLH3 A0A1L8DDT3 A0A067R4F2 A0A1B6FU38 A0A0T6B8V2 A0A1B6D8K7 T1HK07 A0A1B6ECV3 A0A232F0V2 A0A2M4A3J7 K7IXB7 A0A0P5D418 A0A0P5DB75 A0A0N8DVF3 A0A0P5YYJ2 A0A0P5A1H5 A0A182X4Y8 E9FWB8 A0A2H8TYW2 J9K3H1 A0A2J7RKG8 A0A087UQP2 A0A0P6IHC9 A0A0P6GFU0
Ontologies
Topology
Subcellular location
Membrane
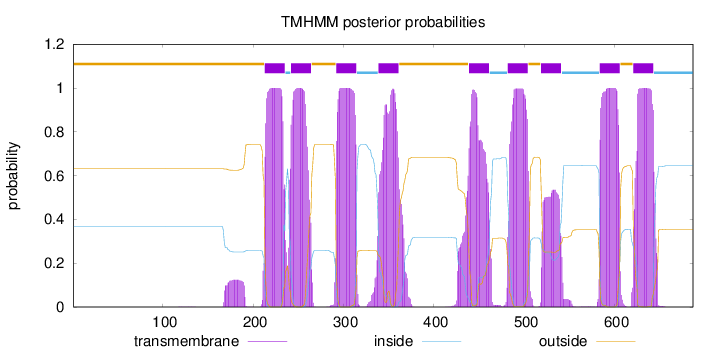
Length:
687
Number of predicted TMHs:
9
Exp number of AAs in TMHs:
196.30286
Exp number, first 60 AAs:
0.00041
Total prob of N-in:
0.36879
outside
1 - 212
TMhelix
213 - 235
inside
236 - 241
TMhelix
242 - 264
outside
265 - 291
TMhelix
292 - 314
inside
315 - 338
TMhelix
339 - 361
outside
362 - 438
TMhelix
439 - 461
inside
462 - 481
TMhelix
482 - 504
outside
505 - 518
TMhelix
519 - 541
inside
542 - 583
TMhelix
584 - 606
outside
607 - 620
TMhelix
621 - 643
inside
644 - 687
Population Genetic Test Statistics
Pi
184.828118
Theta
168.632499
Tajima's D
-0.777718
CLR
0.587546
CSRT
0.181640917954102
Interpretation
Possibly Positive selection
