Gene
KWMTBOMO08798
Annotation
PREDICTED:_torsin-like_protein_isoform_X2_[Bombyx_mori]
Full name
Torsin
+ More
Torsin-like protein
Torsin-like protein
Location in the cell
Cytoplasmic Reliability : 1.88
Sequence
CDS
ATGTTTTTAAGTAGCACGTTATCTGAACCAATCACGATATCTTTAGTTGGATCAGCTATCGTTCTTGCTAGCGGATGGTATAAATGGGATACTTTAAAGGATGCCACATATTGTAAATTTACAGAATGTTGTAATGATTATCACATACCTTTTGATGTACAAAAATTGAGAGATTCTCTTTCACAGCGAATGTTTGGTCAGCCACTGGTCAATGAACTTTTCAATATTATCTCAGCTCACAAAGAGAATATTAATGAAAGCAATGGGAACAAGAAAGCTCTAGTAATAAGTCTCCATGGCTGGTCAGGTGTTGGTAAAAATTTTGCCTCTACAATGATTGCCGAAGCTATCTATCGCAAAGGCATGCAGAGCAACTATGTAAAACTATTTATGGGTAAAAAAGATTTCGATTGCTATGAATTGGAAAAGAAAAAACAAATGTTAGTTAATACATTGAACACTCTGGTGAAGAAATGTCCCAAATCTCTGATTATCTTTGATGAGATACATCACATGTGTCCATCAGTGCTTGATACAATAATACCAATGCTCGATCACCACAGTGCTGTTGATGAGGTGGATTATAGAAACACCATCTTTATTTTTATATCAAACATAGGAGGACATGAAATAGCAACAAAGCTTTTAGAACTCTACGAAAAAGGTGTAACTCGATCAGAAGTTGAATTCCGTGATTTTGAACCTATCATTCGGAGGACAGCATATTTCCAAGGTGGCTTCGAGAAAGCGGCCGCCATTGCACACCATTTGATCGATCACTATGTACCTTTTTTACCCTTGGAACAACATCACGTCGAAATGTGTGCACTTGCAGAATTTAAAGCTAACGGCGTTGAAAAGCCGTCGAAGAAAATGATGGCGGATGCCTTAACTGTTATAACATACGGTCCGCCAGAGACTCAACCTATTTTTGCAAATAATGGATGCAGACGATTCACAAGACACATTCCATATGTAATAAAAAAGCACACTAGAAAAACAGAATTATGA
Protein
MFLSSTLSEPITISLVGSAIVLASGWYKWDTLKDATYCKFTECCNDYHIPFDVQKLRDSLSQRMFGQPLVNELFNIISAHKENINESNGNKKALVISLHGWSGVGKNFASTMIAEAIYRKGMQSNYVKLFMGKKDFDCYELEKKKQMLVNTLNTLVKKCPKSLIIFDEIHHMCPSVLDTIIPMLDHHSAVDEVDYRNTIFIFISNIGGHEIATKLLELYEKGVTRSEVEFRDFEPIIRRTAYFQGGFEKAAAIAHHLIDHYVPFLPLEQHHVEMCALAEFKANGVEKPSKKMMADALTVITYGPPETQPIFANNGCRRFTRHIPYVIKKHTRKTEL
Summary
Description
May serve as a molecular chaperone assisting in the proper folding of secreted and/or membrane proteins.
Similarity
Belongs to the ClpA/ClpB family. Torsin subfamily.
Keywords
ATP-binding
Chaperone
Complete proteome
Endoplasmic reticulum
Glycoprotein
Nucleotide-binding
Polymorphism
Reference proteome
Signal
Feature
chain Torsin
Uniprot
A0A2H1W0J6
A0A194PWJ8
A0A194QT35
S4PHJ3
A0A0L7LG55
A0A437AYQ7
+ More
A0A212F0X6 A0A2A4K4K2 Q17CV2 A0A1I8NMZ1 A0A182V258 A0A1L8EAX5 A0A1L8EB05 A0A1L8EAG2 A0A1L8EA57 A0A1L8EAY6 A0A1L8EBD1 A0A1S4H1D8 A0A1A9WIN6 A0A1Q3G2P5 T1P875 A0A1I8NII3 A0A034WDE2 A0A2M3ZE57 A0A1B0FIY7 A0A2M3ZAR6 A0A2M4BS86 A0A1Y1MPI9 W8CDH5 A0A0K8UZI2 Q7QG78 F6JQ92 F6JQ98 F6JQA7 A0A1A9ZEK9 B4Q036 A0A1A9XBS4 F6JQA6 O77277 F6JQ95 A0A023EQ48 A0A1A9VCM7 A0A182K169 A0A1W7R7Z2 A0A1B0BDX6 W5J3N3 F6JQA2 A0A0L0C891 A0A0A1WG10 A0A131YTI2 A0A0V1MW92 B4L707 A0A224ZAJ0 A0A0V1IWP1 A0A0V0WY85 T1E8F4 A0A1J1ITS0 A0A1W4UII9 A0A0V1CSA1 L7MFB9 A0A336K870 A0A0K8U3J1 D6W8J5 A0A131XJ30 A0A195E750 A0A0N4UMN3 A0A1I7VNH5 B3NU51 V9L297 A0A0P4WLB8 A0A182VV27 A0A0V0U157 A0A1S0U7L7 A0A0V1FEP2 A0A026W368 A0A0V1MWS3 A0A023G8U9 A0A0M4FA77 G3MH46 B4N290 B4R4P3 A0A293N2U0 B4I0W8 B4MA82 B7PA06 E9IUC7 A0A158NGJ6 A0A0V1E5B9 V9KVY4 A0A182E220 A0A2K6VGX0 A0A1I8EPX3 A0A1L8DUT1 A0A3B0JQF0 Q29G80 E2A8N8 A0A0K8RDV7 A0A0H5SHH5 A0A195B7Z3 A0A2J7QV80
A0A212F0X6 A0A2A4K4K2 Q17CV2 A0A1I8NMZ1 A0A182V258 A0A1L8EAX5 A0A1L8EB05 A0A1L8EAG2 A0A1L8EA57 A0A1L8EAY6 A0A1L8EBD1 A0A1S4H1D8 A0A1A9WIN6 A0A1Q3G2P5 T1P875 A0A1I8NII3 A0A034WDE2 A0A2M3ZE57 A0A1B0FIY7 A0A2M3ZAR6 A0A2M4BS86 A0A1Y1MPI9 W8CDH5 A0A0K8UZI2 Q7QG78 F6JQ92 F6JQ98 F6JQA7 A0A1A9ZEK9 B4Q036 A0A1A9XBS4 F6JQA6 O77277 F6JQ95 A0A023EQ48 A0A1A9VCM7 A0A182K169 A0A1W7R7Z2 A0A1B0BDX6 W5J3N3 F6JQA2 A0A0L0C891 A0A0A1WG10 A0A131YTI2 A0A0V1MW92 B4L707 A0A224ZAJ0 A0A0V1IWP1 A0A0V0WY85 T1E8F4 A0A1J1ITS0 A0A1W4UII9 A0A0V1CSA1 L7MFB9 A0A336K870 A0A0K8U3J1 D6W8J5 A0A131XJ30 A0A195E750 A0A0N4UMN3 A0A1I7VNH5 B3NU51 V9L297 A0A0P4WLB8 A0A182VV27 A0A0V0U157 A0A1S0U7L7 A0A0V1FEP2 A0A026W368 A0A0V1MWS3 A0A023G8U9 A0A0M4FA77 G3MH46 B4N290 B4R4P3 A0A293N2U0 B4I0W8 B4MA82 B7PA06 E9IUC7 A0A158NGJ6 A0A0V1E5B9 V9KVY4 A0A182E220 A0A2K6VGX0 A0A1I8EPX3 A0A1L8DUT1 A0A3B0JQF0 Q29G80 E2A8N8 A0A0K8RDV7 A0A0H5SHH5 A0A195B7Z3 A0A2J7QV80
Pubmed
26354079
23622113
26227816
22118469
17510324
12364791
+ More
25315136 25348373 28004739 24495485 21173424 21383965 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18477586 19126864 10731137 12537569 24945155 20920257 23761445 26108605 25830018 26830274 28797301 25576852 18362917 19820115 28049606 25217238 24402279 24508170 30249741 22216098 21282665 21347285 22919073 26850696 15632085 20798317 17885136
25315136 25348373 28004739 24495485 21173424 21383965 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18477586 19126864 10731137 12537569 24945155 20920257 23761445 26108605 25830018 26830274 28797301 25576852 18362917 19820115 28049606 25217238 24402279 24508170 30249741 22216098 21282665 21347285 22919073 26850696 15632085 20798317 17885136
EMBL
ODYU01005599
SOQ46601.1
KQ459596
KPI95510.1
KQ461195
KPJ06701.1
+ More
GAIX01000454 JAA92106.1 JTDY01001240 KOB74437.1 RSAL01000266 RVE43254.1 AGBW02011012 OWR47382.1 NWSH01000179 PCG78693.1 CH477304 EAT44169.1 GFDG01002879 JAV15920.1 GFDG01002881 JAV15918.1 GFDG01003190 JAV15609.1 GFDG01003191 JAV15608.1 GFDG01002882 JAV15917.1 GFDG01002880 JAV15919.1 AAAB01008839 GFDL01000975 JAV34070.1 KA644792 AFP59421.1 GAKP01005351 JAC53601.1 GGFM01006075 MBW26826.1 CCAG010017454 GGFM01004862 MBW25613.1 GGFJ01006806 MBW55947.1 GEZM01025083 JAV87599.1 GAMC01001936 JAC04620.1 GDHF01020298 JAI32016.1 EAA05928.4 GQ326645 GQ326646 GQ326647 GQ326649 GQ326651 GQ326656 GQ326659 ACU26618.1 ACU26619.1 ACU26620.1 ACU26622.1 ACU26624.1 ACU26629.1 ACU26632.1 GQ326648 GQ326652 GQ326653 GQ326654 GQ326655 GQ326658 GQ326663 ACU26621.1 ACU26625.1 ACU26626.1 ACU26627.1 ACU26628.1 ACU26631.1 ACU26636.1 GQ326662 ACU26635.1 CM000162 EDX01188.1 GQ326660 GQ326661 AE014298 ACU26633.1 ACU26634.1 AHN59337.1 AM999060 AM999061 AM999062 AM999063 AM999064 AM999065 AM999066 AM999067 AM999068 AM999069 FM246173 FM246174 FM246175 FM246176 FM246177 FM246178 FM246179 FM246180 FM246181 FM246182 FM246183 FM246184 AF236156 AL031766 AY069592 BT003553 GQ326650 GQ326664 ACU26623.1 ACU26637.1 GAPW01002423 JAC11175.1 GEHC01000389 JAV47256.1 JXJN01012727 ADMH02002133 ETN58446.1 GQ326657 ACU26630.1 JRES01000767 KNC28465.1 GBXI01016974 JAC97317.1 GEDV01006290 JAP82267.1 JYDO01000032 KRZ76027.1 CH933812 EDW06153.2 GFPF01012536 MAA23682.1 JYDS01000074 KRZ27178.1 JYDK01000038 KRX80748.1 GAMD01002227 JAA99363.1 CVRI01000057 CRL01913.1 JYDI01000110 KRY52161.1 GACK01002128 JAA62906.1 UFQS01000118 UFQT01000118 SSX00022.1 SSX20402.1 GDHF01031045 JAI21269.1 KQ971312 EEZ98367.1 GEFH01002396 JAP66185.1 KQ979568 KYN21025.1 UYYG01001229 VDN60649.1 CH954180 EDV45827.1 JW872880 AFP05398.1 GDRN01039269 JAI67199.1 JYDJ01000084 KRX45007.1 JH712075 EFO26311.1 JYDT01000114 KRY84418.1 KK107487 QOIP01000006 EZA50031.1 RLU21666.1 KRZ76028.1 GBBM01005234 JAC30184.1 CP012528 ALC49600.1 JO841197 AEO32814.1 CH963925 EDW78479.1 CM000366 EDX17090.1 GFWV01022656 MAA47383.1 CH480819 EDW53149.1 CH940655 EDW66141.2 ABJB010030699 ABJB010242574 ABJB010333825 ABJB010377240 DS668357 EEC03428.1 GL765940 EFZ15825.1 ADTU01015037 JYDR01000100 KRY68971.1 JW869937 AFP02455.1 UYRW01000287 VDK65395.1 CMVM020000346 GFDF01004049 JAV10035.1 OUUW01000003 SPP77680.1 CH379064 EAL32230.3 GL437652 EFN70225.1 GADI01004456 JAA69352.1 LN856856 CRZ23294.1 KQ976574 KYM80362.1 NEVH01010478 PNF32486.1
GAIX01000454 JAA92106.1 JTDY01001240 KOB74437.1 RSAL01000266 RVE43254.1 AGBW02011012 OWR47382.1 NWSH01000179 PCG78693.1 CH477304 EAT44169.1 GFDG01002879 JAV15920.1 GFDG01002881 JAV15918.1 GFDG01003190 JAV15609.1 GFDG01003191 JAV15608.1 GFDG01002882 JAV15917.1 GFDG01002880 JAV15919.1 AAAB01008839 GFDL01000975 JAV34070.1 KA644792 AFP59421.1 GAKP01005351 JAC53601.1 GGFM01006075 MBW26826.1 CCAG010017454 GGFM01004862 MBW25613.1 GGFJ01006806 MBW55947.1 GEZM01025083 JAV87599.1 GAMC01001936 JAC04620.1 GDHF01020298 JAI32016.1 EAA05928.4 GQ326645 GQ326646 GQ326647 GQ326649 GQ326651 GQ326656 GQ326659 ACU26618.1 ACU26619.1 ACU26620.1 ACU26622.1 ACU26624.1 ACU26629.1 ACU26632.1 GQ326648 GQ326652 GQ326653 GQ326654 GQ326655 GQ326658 GQ326663 ACU26621.1 ACU26625.1 ACU26626.1 ACU26627.1 ACU26628.1 ACU26631.1 ACU26636.1 GQ326662 ACU26635.1 CM000162 EDX01188.1 GQ326660 GQ326661 AE014298 ACU26633.1 ACU26634.1 AHN59337.1 AM999060 AM999061 AM999062 AM999063 AM999064 AM999065 AM999066 AM999067 AM999068 AM999069 FM246173 FM246174 FM246175 FM246176 FM246177 FM246178 FM246179 FM246180 FM246181 FM246182 FM246183 FM246184 AF236156 AL031766 AY069592 BT003553 GQ326650 GQ326664 ACU26623.1 ACU26637.1 GAPW01002423 JAC11175.1 GEHC01000389 JAV47256.1 JXJN01012727 ADMH02002133 ETN58446.1 GQ326657 ACU26630.1 JRES01000767 KNC28465.1 GBXI01016974 JAC97317.1 GEDV01006290 JAP82267.1 JYDO01000032 KRZ76027.1 CH933812 EDW06153.2 GFPF01012536 MAA23682.1 JYDS01000074 KRZ27178.1 JYDK01000038 KRX80748.1 GAMD01002227 JAA99363.1 CVRI01000057 CRL01913.1 JYDI01000110 KRY52161.1 GACK01002128 JAA62906.1 UFQS01000118 UFQT01000118 SSX00022.1 SSX20402.1 GDHF01031045 JAI21269.1 KQ971312 EEZ98367.1 GEFH01002396 JAP66185.1 KQ979568 KYN21025.1 UYYG01001229 VDN60649.1 CH954180 EDV45827.1 JW872880 AFP05398.1 GDRN01039269 JAI67199.1 JYDJ01000084 KRX45007.1 JH712075 EFO26311.1 JYDT01000114 KRY84418.1 KK107487 QOIP01000006 EZA50031.1 RLU21666.1 KRZ76028.1 GBBM01005234 JAC30184.1 CP012528 ALC49600.1 JO841197 AEO32814.1 CH963925 EDW78479.1 CM000366 EDX17090.1 GFWV01022656 MAA47383.1 CH480819 EDW53149.1 CH940655 EDW66141.2 ABJB010030699 ABJB010242574 ABJB010333825 ABJB010377240 DS668357 EEC03428.1 GL765940 EFZ15825.1 ADTU01015037 JYDR01000100 KRY68971.1 JW869937 AFP02455.1 UYRW01000287 VDK65395.1 CMVM020000346 GFDF01004049 JAV10035.1 OUUW01000003 SPP77680.1 CH379064 EAL32230.3 GL437652 EFN70225.1 GADI01004456 JAA69352.1 LN856856 CRZ23294.1 KQ976574 KYM80362.1 NEVH01010478 PNF32486.1
Proteomes
UP000053268
UP000053240
UP000037510
UP000283053
UP000007151
UP000218220
+ More
UP000008820 UP000095300 UP000075903 UP000091820 UP000095301 UP000092444 UP000007062 UP000092445 UP000002282 UP000092443 UP000000803 UP000078200 UP000075881 UP000092460 UP000000673 UP000037069 UP000054843 UP000009192 UP000054805 UP000054673 UP000183832 UP000192221 UP000054653 UP000007266 UP000078492 UP000038040 UP000274756 UP000095285 UP000008711 UP000075920 UP000055048 UP000054995 UP000053097 UP000279307 UP000092553 UP000007798 UP000000304 UP000001292 UP000008792 UP000001555 UP000005205 UP000054632 UP000077448 UP000271087 UP000024404 UP000093561 UP000268350 UP000001819 UP000000311 UP000006672 UP000078540 UP000235965
UP000008820 UP000095300 UP000075903 UP000091820 UP000095301 UP000092444 UP000007062 UP000092445 UP000002282 UP000092443 UP000000803 UP000078200 UP000075881 UP000092460 UP000000673 UP000037069 UP000054843 UP000009192 UP000054805 UP000054673 UP000183832 UP000192221 UP000054653 UP000007266 UP000078492 UP000038040 UP000274756 UP000095285 UP000008711 UP000075920 UP000055048 UP000054995 UP000053097 UP000279307 UP000092553 UP000007798 UP000000304 UP000001292 UP000008792 UP000001555 UP000005205 UP000054632 UP000077448 UP000271087 UP000024404 UP000093561 UP000268350 UP000001819 UP000000311 UP000006672 UP000078540 UP000235965
Pfam
Interpro
IPR027417
P-loop_NTPase
+ More
IPR010448 Torsin
IPR003593 AAA+_ATPase
IPR001270 ClpA/B
IPR017378 Torsin_1/2
IPR030553 Torsin-like
IPR003960 ATPase_AAA_CS
IPR003859 Galactosyl_T
IPR029044 Nucleotide-diphossugar_trans
IPR003959 ATPase_AAA_core
IPR027995 Galactosyl_T_N
IPR027791 Galactosyl_T_C
IPR041569 AAA_lid_3
IPR030549 Torsin-1A
IPR013924 RNase_H2_suC
IPR010448 Torsin
IPR003593 AAA+_ATPase
IPR001270 ClpA/B
IPR017378 Torsin_1/2
IPR030553 Torsin-like
IPR003960 ATPase_AAA_CS
IPR003859 Galactosyl_T
IPR029044 Nucleotide-diphossugar_trans
IPR003959 ATPase_AAA_core
IPR027995 Galactosyl_T_N
IPR027791 Galactosyl_T_C
IPR041569 AAA_lid_3
IPR030549 Torsin-1A
IPR013924 RNase_H2_suC
Gene 3D
ProteinModelPortal
A0A2H1W0J6
A0A194PWJ8
A0A194QT35
S4PHJ3
A0A0L7LG55
A0A437AYQ7
+ More
A0A212F0X6 A0A2A4K4K2 Q17CV2 A0A1I8NMZ1 A0A182V258 A0A1L8EAX5 A0A1L8EB05 A0A1L8EAG2 A0A1L8EA57 A0A1L8EAY6 A0A1L8EBD1 A0A1S4H1D8 A0A1A9WIN6 A0A1Q3G2P5 T1P875 A0A1I8NII3 A0A034WDE2 A0A2M3ZE57 A0A1B0FIY7 A0A2M3ZAR6 A0A2M4BS86 A0A1Y1MPI9 W8CDH5 A0A0K8UZI2 Q7QG78 F6JQ92 F6JQ98 F6JQA7 A0A1A9ZEK9 B4Q036 A0A1A9XBS4 F6JQA6 O77277 F6JQ95 A0A023EQ48 A0A1A9VCM7 A0A182K169 A0A1W7R7Z2 A0A1B0BDX6 W5J3N3 F6JQA2 A0A0L0C891 A0A0A1WG10 A0A131YTI2 A0A0V1MW92 B4L707 A0A224ZAJ0 A0A0V1IWP1 A0A0V0WY85 T1E8F4 A0A1J1ITS0 A0A1W4UII9 A0A0V1CSA1 L7MFB9 A0A336K870 A0A0K8U3J1 D6W8J5 A0A131XJ30 A0A195E750 A0A0N4UMN3 A0A1I7VNH5 B3NU51 V9L297 A0A0P4WLB8 A0A182VV27 A0A0V0U157 A0A1S0U7L7 A0A0V1FEP2 A0A026W368 A0A0V1MWS3 A0A023G8U9 A0A0M4FA77 G3MH46 B4N290 B4R4P3 A0A293N2U0 B4I0W8 B4MA82 B7PA06 E9IUC7 A0A158NGJ6 A0A0V1E5B9 V9KVY4 A0A182E220 A0A2K6VGX0 A0A1I8EPX3 A0A1L8DUT1 A0A3B0JQF0 Q29G80 E2A8N8 A0A0K8RDV7 A0A0H5SHH5 A0A195B7Z3 A0A2J7QV80
A0A212F0X6 A0A2A4K4K2 Q17CV2 A0A1I8NMZ1 A0A182V258 A0A1L8EAX5 A0A1L8EB05 A0A1L8EAG2 A0A1L8EA57 A0A1L8EAY6 A0A1L8EBD1 A0A1S4H1D8 A0A1A9WIN6 A0A1Q3G2P5 T1P875 A0A1I8NII3 A0A034WDE2 A0A2M3ZE57 A0A1B0FIY7 A0A2M3ZAR6 A0A2M4BS86 A0A1Y1MPI9 W8CDH5 A0A0K8UZI2 Q7QG78 F6JQ92 F6JQ98 F6JQA7 A0A1A9ZEK9 B4Q036 A0A1A9XBS4 F6JQA6 O77277 F6JQ95 A0A023EQ48 A0A1A9VCM7 A0A182K169 A0A1W7R7Z2 A0A1B0BDX6 W5J3N3 F6JQA2 A0A0L0C891 A0A0A1WG10 A0A131YTI2 A0A0V1MW92 B4L707 A0A224ZAJ0 A0A0V1IWP1 A0A0V0WY85 T1E8F4 A0A1J1ITS0 A0A1W4UII9 A0A0V1CSA1 L7MFB9 A0A336K870 A0A0K8U3J1 D6W8J5 A0A131XJ30 A0A195E750 A0A0N4UMN3 A0A1I7VNH5 B3NU51 V9L297 A0A0P4WLB8 A0A182VV27 A0A0V0U157 A0A1S0U7L7 A0A0V1FEP2 A0A026W368 A0A0V1MWS3 A0A023G8U9 A0A0M4FA77 G3MH46 B4N290 B4R4P3 A0A293N2U0 B4I0W8 B4MA82 B7PA06 E9IUC7 A0A158NGJ6 A0A0V1E5B9 V9KVY4 A0A182E220 A0A2K6VGX0 A0A1I8EPX3 A0A1L8DUT1 A0A3B0JQF0 Q29G80 E2A8N8 A0A0K8RDV7 A0A0H5SHH5 A0A195B7Z3 A0A2J7QV80
PDB
5J1S
E-value=3.68966e-38,
Score=396
Ontologies
GO
GO:0005524
GO:0016021
GO:0005788
GO:0051085
GO:0005635
GO:0005783
GO:0071218
GO:0016887
GO:0046467
GO:0043104
GO:0040011
GO:0030307
GO:1903741
GO:0048085
GO:0007504
GO:0071426
GO:0042053
GO:0008057
GO:0070328
GO:0010883
GO:0070050
GO:0044255
GO:0005975
GO:0016757
GO:0045104
GO:0031175
GO:0072321
GO:0006998
GO:0007155
GO:0048489
GO:0045202
GO:0008092
GO:0051082
GO:0006401
GO:0032299
GO:0005622
GO:0005840
GO:0006412
GO:0008097
GO:0016817
GO:0008270
GO:0043565
GO:0016876
GO:0043039
PANTHER
Topology
Subcellular location
Endoplasmic reticulum lumen
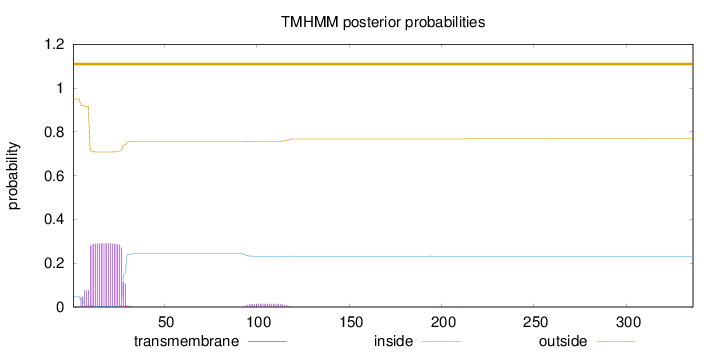
Length:
336
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
6.06515
Exp number, first 60 AAs:
5.74322
Total prob of N-in:
0.04948
outside
1 - 336
Population Genetic Test Statistics
Pi
204.251105
Theta
233.320918
Tajima's D
-1.850012
CLR
1856.882961
CSRT
0.0266986650667467
Interpretation
Uncertain
