Gene
KWMTBOMO08792
Annotation
PREDICTED:_protein-associating_with_the_carboxyl-terminal_domain_of_ezrin_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.802 Nuclear Reliability : 2.066
Sequence
CDS
ATGGGTAACGAAAACTCACAGCTAAGTGGCTTGGAAATAGAGGATAAAGCTGTAGAGATAACAGAATTTTGGCAACAATATCAAGCCACTATAAAAGATAGTTGTAGATACTTCAGTCTGAAAGGGGATGGTTCAGTGTCTGTTTTCAAAGGAGAAGCGGCTGCAGGACCACTTTGGGCTGTATCGACACCTCTAGAAAAATTTTGTAATAATTTATTAAAATATCGACATCCATGTATTGTGAGATACATATCAGCATGGCAACAGCGTTCGACACTGCATTTGGCAACAGAATATGTGCTACCTCTAAGTCATGTTCTGAAAACTCTTTCACCTTTACAAATTTGCATTGGATTAAACAACATTCTTAGAGCTTTAGTATTCTTGCATGATCAGGCTAGCATGTCACATAATAATGTCAGTACTTCAGCTGTATATGTGACAGGTGATGGCCAGTGGAAACTAGGTGGCTTGCAATACTTATGTCCATTTTCTGAGCTCACACCGGCTTATTTAAAACATGCTAGAATCCATAGGTATGATAAAGCCATTGATCCCAATGAAGATTCAAATGAGCTAACAACAAAAGTGGATCAATATGGTTTTGCAGTACTTGTTGAAGATGTTCTAAAGGGACAAAGCGATGATGAGGTACCATATTTAACAAATTTTAAAAAATACTGTAAAGACAACTTGAAAAATGAAGATCCAACAAAAAGGTCAAAGCTATCAGAGGTTTTGCAACACAATTTTTTCAACCACACATTTATCAGTATTTATAAGTTTCTTACATTCCTGCCTTTGAAAAGCGAAGATGAAAGAAGTGAATTCTTCACAACCATTTTAAACAATTTGAAATGTTATGATGAGGAAACTGTAGCTAAGCAATTGAGTGGATTGTTGCTTTCAAGACTCACAATGTTAGATGCAACTGCACGGAGGGAAGTCATTCCATATATCCTCGAACCTTGTAATGAGAGACAACAAAATAGTGAACATCCAGGTTTTTTTAGTTTAAACGTGTTCAAAGAGCATGTGAAACCTAGACTTTTGCAACTGTTCAGTGTCAGAGACAGTCAGATAAGAATGTTGTTATTGATGCATTTTTCAAAGTATATTCATGTCTTCACACACGAGGAGCTCTCGCAACATATACTACCCGAATTACTGTTGGGTATAAAAGACACAGATGATAACTTAGTAGCGTCGACATTAATATGCCTAAGTGTCTTAGTACCTATACTCGGAGCTGCAGCCGTTGTAGGCGGAAAGAGATCAAAATATTTTACTGACGGAAGACCAAACTGTAAAAACAACTTGATAAAATCAACAAGCATCAACAAGCAAGATAAAAAAGTGTATCTCTTAGAAAAATCATTAAGGGAATTACCCTGTTCAGAGGATGTTTATGAAAATGACAATACCGGTCTGACCACTTTTACGAATATAAACTTAAATGAAAGACCTACACCAGTCGGCGGTGAATCTTTGGAGGAAGAAGTAATATTAAGAGAATCTACAATTAATCAAGTTAACAGCGAAGAGGATTGGAGTGATTGGGATAACACTAACCGTCAGCTCACTATGGCAGAAACAGAAAGCATCTCGATTGAAGCAGATACCATTTATTTGAATACCGAAAAAAAAGAAGTACCCGTAATCGAAACGAACGTCGCAGCAGAAGATTCGGCATTAAAAAAATCATTGCTTCAAAAAGCAGCCATTGATGCCAAAAGAAACATTGTAGACATATCAGAGTTGGATATTAAACATCAAAAAACAAGTAATACAAAAAAAGGTGCTGACGAATTTGACTTTTTCGCTGACATGAAGCCTGTTATCGAGAAGGCCGCTGTTGTCAGTATCGATGATTTGGTTTCGAAAGAATTTAGCAGCAAGTTAACGTTCGTACCCGATGAAAACCAAGACGAGAACGAAGGTTGGGGTGAAAACTGGAATGATTAA
Protein
MGNENSQLSGLEIEDKAVEITEFWQQYQATIKDSCRYFSLKGDGSVSVFKGEAAAGPLWAVSTPLEKFCNNLLKYRHPCIVRYISAWQQRSTLHLATEYVLPLSHVLKTLSPLQICIGLNNILRALVFLHDQASMSHNNVSTSAVYVTGDGQWKLGGLQYLCPFSELTPAYLKHARIHRYDKAIDPNEDSNELTTKVDQYGFAVLVEDVLKGQSDDEVPYLTNFKKYCKDNLKNEDPTKRSKLSEVLQHNFFNHTFISIYKFLTFLPLKSEDERSEFFTTILNNLKCYDEETVAKQLSGLLLSRLTMLDATARREVIPYILEPCNERQQNSEHPGFFSLNVFKEHVKPRLLQLFSVRDSQIRMLLLMHFSKYIHVFTHEELSQHILPELLLGIKDTDDNLVASTLICLSVLVPILGAAAVVGGKRSKYFTDGRPNCKNNLIKSTSINKQDKKVYLLEKSLRELPCSEDVYENDNTGLTTFTNINLNERPTPVGGESLEEEVILRESTINQVNSEEDWSDWDNTNRQLTMAETESISIEADTIYLNTEKKEVPVIETNVAAEDSALKKSLLQKAAIDAKRNIVDISELDIKHQKTSNTKKGADEFDFFADMKPVIEKAAVVSIDDLVSKEFSSKLTFVPDENQDENEGWGENWND
Summary
Uniprot
A0A1E1WUI8
A0A2H1W0Q4
A0A2A4K6I2
A0A3S2L157
A0A212F0Y8
A0A194PRX7
+ More
A0A194QNZ3 A0A1E1W001 E2BM79 A0A026WWV8 A0A151X632 A0A195F941 A0A2J7QXC4 A0A158NCL4 A0A195BYI6 E2AK74 F4WFF4 A0A0L7R4Y9 D6X3F4 A0A154P9C7 E9IMB6 A0A310SBM0 A0A088AL07 A0A067QSZ0 A0A0M8ZQ53 A0A2A3E312 K7JLJ8 A0A0C9PIX5 A0A232FLF2 A0A1Y1MEU6 A0A2P8Z0G7 A0A195CQH2 E0VTE2 A0A1B6DLW2 A0A182IMA6 A0A182YP63 A0A182KYK2 A0A182TFQ8 A0A182VSI7 A0A182X891 A0A182NBC8 Q7PWA7 A0A182USK2 A0A182JQD9 A0A2R7WP49 A0A182HZ27 A0A2M4ADE0 A0A182LZW0 A0A182QDQ3 N6TPH8 A0A182F410 A0A1B6LET5 A0A182RWE3 A0A2M3Z583 A0A182SCH3 W5JR81 A0A195ELX5 A0A2M4BG55 A0A2M3Z557 A0A2M3ZH58 A0A2M4BG21 A0A1B6H1U6 A0A1B6GZJ3 A0A182PU30 A0A084VT33 A0A182G2C1 U5EYE9 Q16SM6 A0A2M4BI01 Q17G01 A0A1S4F467 A0A0P4VW33 B0X7P1 A0A0A9Y667 A0A1L8DLL7 A0A1L8DLL5 B4LQR1 A0A0M4ESP8 A0A336M3N7 A0A3B0JTU2 A0A1A9ZW09 A0A1B0CFI3 A0A1I8PMZ2 B4KFU3 B4GXC5 A0A1W4W552 T1PCR0 A0A034W1B6 A0A1W4W9F6
A0A194QNZ3 A0A1E1W001 E2BM79 A0A026WWV8 A0A151X632 A0A195F941 A0A2J7QXC4 A0A158NCL4 A0A195BYI6 E2AK74 F4WFF4 A0A0L7R4Y9 D6X3F4 A0A154P9C7 E9IMB6 A0A310SBM0 A0A088AL07 A0A067QSZ0 A0A0M8ZQ53 A0A2A3E312 K7JLJ8 A0A0C9PIX5 A0A232FLF2 A0A1Y1MEU6 A0A2P8Z0G7 A0A195CQH2 E0VTE2 A0A1B6DLW2 A0A182IMA6 A0A182YP63 A0A182KYK2 A0A182TFQ8 A0A182VSI7 A0A182X891 A0A182NBC8 Q7PWA7 A0A182USK2 A0A182JQD9 A0A2R7WP49 A0A182HZ27 A0A2M4ADE0 A0A182LZW0 A0A182QDQ3 N6TPH8 A0A182F410 A0A1B6LET5 A0A182RWE3 A0A2M3Z583 A0A182SCH3 W5JR81 A0A195ELX5 A0A2M4BG55 A0A2M3Z557 A0A2M3ZH58 A0A2M4BG21 A0A1B6H1U6 A0A1B6GZJ3 A0A182PU30 A0A084VT33 A0A182G2C1 U5EYE9 Q16SM6 A0A2M4BI01 Q17G01 A0A1S4F467 A0A0P4VW33 B0X7P1 A0A0A9Y667 A0A1L8DLL7 A0A1L8DLL5 B4LQR1 A0A0M4ESP8 A0A336M3N7 A0A3B0JTU2 A0A1A9ZW09 A0A1B0CFI3 A0A1I8PMZ2 B4KFU3 B4GXC5 A0A1W4W552 T1PCR0 A0A034W1B6 A0A1W4W9F6
Pubmed
EMBL
GDQN01000420
JAT90634.1
ODYU01005599
SOQ46607.1
NWSH01000105
PCG79514.1
+ More
RSAL01000268 RVE43213.1 AGBW02011012 OWR47387.1 KQ459596 KPI95504.1 KQ461195 KPJ06695.1 GDQN01010739 JAT80315.1 GL449158 EFN83205.1 KK107077 QOIP01000010 EZA60560.1 RLU17330.1 KQ982490 KYQ55802.1 KQ981727 KYN36958.1 NEVH01009381 PNF33237.1 ADTU01011960 KQ976396 KYM93001.1 GL440194 EFN66178.1 GL888115 EGI67205.1 KQ414654 KOC65908.1 KQ971374 EEZ97411.1 KQ434831 KZC07828.1 GL764129 EFZ18398.1 KQ761783 OAD57040.1 KK853055 KDR11980.1 KQ435989 KOX67696.1 KZ288445 PBC25646.1 GBYB01000918 JAG70685.1 NNAY01000067 OXU31340.1 GEZM01033210 JAV84349.1 PYGN01000255 PSN49978.1 KQ977408 KYN02956.1 DS235762 EEB16648.1 GEDC01010660 JAS26638.1 AAAB01008984 EAA14766.4 KK855200 PTY21408.1 APCN01005849 GGFK01005485 MBW38806.1 AXCM01002560 AXCN02000626 APGK01056791 KB741277 KB631786 ENN71125.1 ERL86080.1 GEBQ01017804 JAT22173.1 GGFM01002921 MBW23672.1 ADMH02000573 ETN65818.1 KQ978691 KYN29218.1 GGFJ01002866 MBW52007.1 GGFM01002884 MBW23635.1 GGFM01007101 MBW27852.1 GGFJ01002865 MBW52006.1 GECZ01001152 JAS68617.1 GECZ01001935 JAS67834.1 ATLV01016154 KE525057 KFB41127.1 JXUM01139033 KQ568831 KXJ68871.1 GANO01000402 JAB59469.1 CH477671 EAT37469.1 GGFJ01003521 MBW52662.1 CH477267 EAT45469.1 GDKW01000300 JAI56295.1 DS232456 EDS42053.1 GBHO01017021 JAG26583.1 GFDF01006726 JAV07358.1 GFDF01006725 JAV07359.1 CH940649 EDW63445.1 CP012523 ALC40400.1 UFQS01000363 UFQT01000363 SSX03177.1 SSX23543.1 OUUW01000010 SPP85534.1 AJWK01010107 CH933807 EDW12069.1 CH479195 EDW27236.1 KA645920 AFP60549.1 GAKP01011379 JAC47573.1
RSAL01000268 RVE43213.1 AGBW02011012 OWR47387.1 KQ459596 KPI95504.1 KQ461195 KPJ06695.1 GDQN01010739 JAT80315.1 GL449158 EFN83205.1 KK107077 QOIP01000010 EZA60560.1 RLU17330.1 KQ982490 KYQ55802.1 KQ981727 KYN36958.1 NEVH01009381 PNF33237.1 ADTU01011960 KQ976396 KYM93001.1 GL440194 EFN66178.1 GL888115 EGI67205.1 KQ414654 KOC65908.1 KQ971374 EEZ97411.1 KQ434831 KZC07828.1 GL764129 EFZ18398.1 KQ761783 OAD57040.1 KK853055 KDR11980.1 KQ435989 KOX67696.1 KZ288445 PBC25646.1 GBYB01000918 JAG70685.1 NNAY01000067 OXU31340.1 GEZM01033210 JAV84349.1 PYGN01000255 PSN49978.1 KQ977408 KYN02956.1 DS235762 EEB16648.1 GEDC01010660 JAS26638.1 AAAB01008984 EAA14766.4 KK855200 PTY21408.1 APCN01005849 GGFK01005485 MBW38806.1 AXCM01002560 AXCN02000626 APGK01056791 KB741277 KB631786 ENN71125.1 ERL86080.1 GEBQ01017804 JAT22173.1 GGFM01002921 MBW23672.1 ADMH02000573 ETN65818.1 KQ978691 KYN29218.1 GGFJ01002866 MBW52007.1 GGFM01002884 MBW23635.1 GGFM01007101 MBW27852.1 GGFJ01002865 MBW52006.1 GECZ01001152 JAS68617.1 GECZ01001935 JAS67834.1 ATLV01016154 KE525057 KFB41127.1 JXUM01139033 KQ568831 KXJ68871.1 GANO01000402 JAB59469.1 CH477671 EAT37469.1 GGFJ01003521 MBW52662.1 CH477267 EAT45469.1 GDKW01000300 JAI56295.1 DS232456 EDS42053.1 GBHO01017021 JAG26583.1 GFDF01006726 JAV07358.1 GFDF01006725 JAV07359.1 CH940649 EDW63445.1 CP012523 ALC40400.1 UFQS01000363 UFQT01000363 SSX03177.1 SSX23543.1 OUUW01000010 SPP85534.1 AJWK01010107 CH933807 EDW12069.1 CH479195 EDW27236.1 KA645920 AFP60549.1 GAKP01011379 JAC47573.1
Proteomes
UP000218220
UP000283053
UP000007151
UP000053268
UP000053240
UP000008237
+ More
UP000053097 UP000279307 UP000075809 UP000078541 UP000235965 UP000005205 UP000078540 UP000000311 UP000007755 UP000053825 UP000007266 UP000076502 UP000005203 UP000027135 UP000053105 UP000242457 UP000002358 UP000215335 UP000245037 UP000078542 UP000009046 UP000075880 UP000076408 UP000075882 UP000075902 UP000075920 UP000076407 UP000075884 UP000007062 UP000075903 UP000075881 UP000075840 UP000075883 UP000075886 UP000019118 UP000030742 UP000069272 UP000075900 UP000075901 UP000000673 UP000078492 UP000075885 UP000030765 UP000069940 UP000249989 UP000008820 UP000002320 UP000008792 UP000092553 UP000268350 UP000092445 UP000092461 UP000095300 UP000009192 UP000008744 UP000192223 UP000095301 UP000192221
UP000053097 UP000279307 UP000075809 UP000078541 UP000235965 UP000005205 UP000078540 UP000000311 UP000007755 UP000053825 UP000007266 UP000076502 UP000005203 UP000027135 UP000053105 UP000242457 UP000002358 UP000215335 UP000245037 UP000078542 UP000009046 UP000075880 UP000076408 UP000075882 UP000075902 UP000075920 UP000076407 UP000075884 UP000007062 UP000075903 UP000075881 UP000075840 UP000075883 UP000075886 UP000019118 UP000030742 UP000069272 UP000075900 UP000075901 UP000000673 UP000078492 UP000075885 UP000030765 UP000069940 UP000249989 UP000008820 UP000002320 UP000008792 UP000092553 UP000268350 UP000092445 UP000092461 UP000095300 UP000009192 UP000008744 UP000192223 UP000095301 UP000192221
Interpro
Gene 3D
ProteinModelPortal
A0A1E1WUI8
A0A2H1W0Q4
A0A2A4K6I2
A0A3S2L157
A0A212F0Y8
A0A194PRX7
+ More
A0A194QNZ3 A0A1E1W001 E2BM79 A0A026WWV8 A0A151X632 A0A195F941 A0A2J7QXC4 A0A158NCL4 A0A195BYI6 E2AK74 F4WFF4 A0A0L7R4Y9 D6X3F4 A0A154P9C7 E9IMB6 A0A310SBM0 A0A088AL07 A0A067QSZ0 A0A0M8ZQ53 A0A2A3E312 K7JLJ8 A0A0C9PIX5 A0A232FLF2 A0A1Y1MEU6 A0A2P8Z0G7 A0A195CQH2 E0VTE2 A0A1B6DLW2 A0A182IMA6 A0A182YP63 A0A182KYK2 A0A182TFQ8 A0A182VSI7 A0A182X891 A0A182NBC8 Q7PWA7 A0A182USK2 A0A182JQD9 A0A2R7WP49 A0A182HZ27 A0A2M4ADE0 A0A182LZW0 A0A182QDQ3 N6TPH8 A0A182F410 A0A1B6LET5 A0A182RWE3 A0A2M3Z583 A0A182SCH3 W5JR81 A0A195ELX5 A0A2M4BG55 A0A2M3Z557 A0A2M3ZH58 A0A2M4BG21 A0A1B6H1U6 A0A1B6GZJ3 A0A182PU30 A0A084VT33 A0A182G2C1 U5EYE9 Q16SM6 A0A2M4BI01 Q17G01 A0A1S4F467 A0A0P4VW33 B0X7P1 A0A0A9Y667 A0A1L8DLL7 A0A1L8DLL5 B4LQR1 A0A0M4ESP8 A0A336M3N7 A0A3B0JTU2 A0A1A9ZW09 A0A1B0CFI3 A0A1I8PMZ2 B4KFU3 B4GXC5 A0A1W4W552 T1PCR0 A0A034W1B6 A0A1W4W9F6
A0A194QNZ3 A0A1E1W001 E2BM79 A0A026WWV8 A0A151X632 A0A195F941 A0A2J7QXC4 A0A158NCL4 A0A195BYI6 E2AK74 F4WFF4 A0A0L7R4Y9 D6X3F4 A0A154P9C7 E9IMB6 A0A310SBM0 A0A088AL07 A0A067QSZ0 A0A0M8ZQ53 A0A2A3E312 K7JLJ8 A0A0C9PIX5 A0A232FLF2 A0A1Y1MEU6 A0A2P8Z0G7 A0A195CQH2 E0VTE2 A0A1B6DLW2 A0A182IMA6 A0A182YP63 A0A182KYK2 A0A182TFQ8 A0A182VSI7 A0A182X891 A0A182NBC8 Q7PWA7 A0A182USK2 A0A182JQD9 A0A2R7WP49 A0A182HZ27 A0A2M4ADE0 A0A182LZW0 A0A182QDQ3 N6TPH8 A0A182F410 A0A1B6LET5 A0A182RWE3 A0A2M3Z583 A0A182SCH3 W5JR81 A0A195ELX5 A0A2M4BG55 A0A2M3Z557 A0A2M3ZH58 A0A2M4BG21 A0A1B6H1U6 A0A1B6GZJ3 A0A182PU30 A0A084VT33 A0A182G2C1 U5EYE9 Q16SM6 A0A2M4BI01 Q17G01 A0A1S4F467 A0A0P4VW33 B0X7P1 A0A0A9Y667 A0A1L8DLL7 A0A1L8DLL5 B4LQR1 A0A0M4ESP8 A0A336M3N7 A0A3B0JTU2 A0A1A9ZW09 A0A1B0CFI3 A0A1I8PMZ2 B4KFU3 B4GXC5 A0A1W4W552 T1PCR0 A0A034W1B6 A0A1W4W9F6
PDB
3VWA
E-value=2.9353e-06,
Score=124
Ontologies
GO
Topology
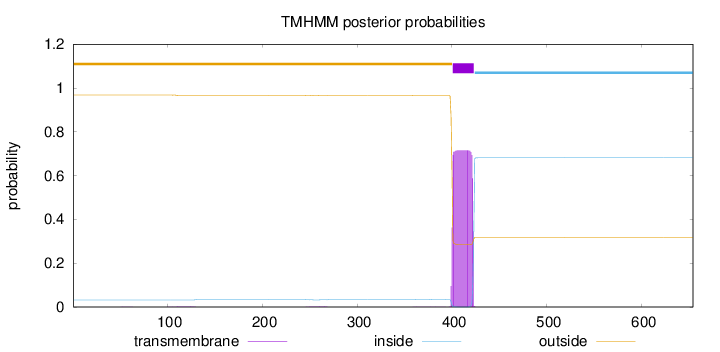
Length:
654
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
16.4631
Exp number, first 60 AAs:
0.00211
Total prob of N-in:
0.03184
outside
1 - 400
TMhelix
401 - 423
inside
424 - 654
Population Genetic Test Statistics
Pi
184.21337
Theta
163.824628
Tajima's D
0.122942
CLR
102.131454
CSRT
0.403129843507825
Interpretation
Possibly Positive selection
