Gene
KWMTBOMO08788
Annotation
PREDICTED:_argininosuccinate_synthase_[Papilio_xuthus]
Full name
Argininosuccinate synthase
Alternative Name
Citrulline--aspartate ligase
Location in the cell
Cytoplasmic Reliability : 3.194
Sequence
CDS
ATGGTTAAAGATCTTGTAATTCTTGCATACTCTGGTGGCTTAGACACTAGTTGTATATTAAAATGGCTAATACAAAAGAACTACGAAGTTGTCTGTTTCATGGCAGATGTCGGACAAAATGAGGACTTCGAAAAAGCTAAAAAGAAAGCTGTAGCAGTTGGTGCCAAAGATGTAGTCGTAGAAGATCTTCGTAAAGAGTTTGTATGTAATTATATGCTGCCTGCAGTACAAATGGGACTTGTCTACGAAAGTCGTTACTACTTGGGAACATCACTTGCCCGACCATGTATAACCGTCGGACTCGTAAACACTGCTAAAAATTTAGGCGCTAAATACATAGCCCACGGAGCCACTGGAAAGGGAAATGACCAAGTGCGGTTTGAATTAAGCATGTATTCGTTATGGCCAGAAGGAAAGATTATAGCACCGTGGCGCATCCCTGAATTCTTCCAAAGGTTTCAGGGAAGAACTGACTTGATTGAGTACGCTAAAAAAGAAAATATACCCGTTTCAGCTACGCCAAAAGAACCTTGGTCCACAGACGAAAATATTATGCATATTAGTTACGAATCTGGGGTATTGGAGGATCCCGCTGCCATTCCACCGGGGGGCATATTTAAAATGACACGAGACTTAGCAAAGGCTGATGATTATGCGAGCGTTTTGGATATACATTTTGAAAAAGGATTGCCTGTTTCTTTACGAATTCCAAGTGGTCACGAAATGGCTCTTCACGTAAACGATCCCCTTGTTATCGTAGAAACGTTAAACAAACTAGGCGGCCAGCATGGGATTGGCAGAATAGACATAGTTGAGAATCGTTTTCTTGGATTGAAGTCCAGAGGTTTGTATGAAACTCCAGCTGGCACCGTATTACATGTGGCGCACGTTGATTTGGAAGCATTCACCCTCGACAGAGAAGTACTGAGAGTGAAAAAATATCTACAAGATAAAATGGCTGATTTTGTATATAACGGATTTTGGTTTTCACCGGAAGCAGCTTTTACAAGGAAATGCCTGGAACTATCCCAGGAGTCTGTTACTGGAACTGTTACCGTTCAGTTATTCAAAGGAAACGTTACCGTTCTGGCAAGAAAAAGTCCAAACAGTTTATACAACAAGGACCTGGTGTCCATGGATATCGCCGGAGGCTTTTCTCCTGAAGATAGTACCGGTTTTATCAATATTAACGCAATTCGCTTGAAGGAATTCGCAAGATTATCCAATCTGAAATAA
Protein
MVKDLVILAYSGGLDTSCILKWLIQKNYEVVCFMADVGQNEDFEKAKKKAVAVGAKDVVVEDLRKEFVCNYMLPAVQMGLVYESRYYLGTSLARPCITVGLVNTAKNLGAKYIAHGATGKGNDQVRFELSMYSLWPEGKIIAPWRIPEFFQRFQGRTDLIEYAKKENIPVSATPKEPWSTDENIMHISYESGVLEDPAAIPPGGIFKMTRDLAKADDYASVLDIHFEKGLPVSLRIPSGHEMALHVNDPLVIVETLNKLGGQHGIGRIDIVENRFLGLKSRGLYETPAGTVLHVAHVDLEAFTLDREVLRVKKYLQDKMADFVYNGFWFSPEAAFTRKCLELSQESVTGTVTVQLFKGNVTVLARKSPNSLYNKDLVSMDIAGGFSPEDSTGFININAIRLKEFARLSNLK
Summary
Description
One of the enzymes of the urea cycle, the metabolic pathway transforming neurotoxic amonia produced by protein catabolism into inocuous urea in the liver of ureotelic animals. Catalyzes the formation of arginosuccinate from aspartate, citrulline and ATP and together with ASL it is responsible for the biosynthesis of arginine in most body tissues.
Catalytic Activity
ATP + L-aspartate + L-citrulline = 2-(N(omega)-L-arginino)succinate + AMP + diphosphate + H(+)
Subunit
Homotetramer.
Similarity
Belongs to the argininosuccinate synthase family.
Belongs to the argininosuccinate synthase family. Type 1 subfamily.
Belongs to the argininosuccinate synthase family. Type 1 subfamily.
Keywords
Amino-acid biosynthesis
Arginine biosynthesis
ATP-binding
Complete proteome
Ligase
Nucleotide-binding
Reference proteome
Urea cycle
Cytoplasm
Phosphoprotein
Feature
chain Argininosuccinate synthase
Uniprot
A0A2A4K5L2
A0A2H1WPY3
A0A212FC20
A0A194QMD4
A0A3S2NBM0
A0A194PPX9
+ More
D6X477 A0A336M4E0 A0A336MHX5 A0A2P8XPL1 A0A2J7QV22 A0A2J7QV24 U5EV09 J3JX12 A0A182SAU0 A0A182K946 A0A067QUJ2 W5J7W2 A0A182N639 A0A182Y4A8 A0A084VRC0 A0A2M4AI34 T1PCL4 A0A1B0CVH8 A0A182P5T4 A0A182RNR2 A0A1I8N6S5 A0A182Q4G0 A0A182VJI6 A0A182WRJ6 A0A182HU80 Q7PR38 A0A182TG76 A0A1I8PQ65 A0A182W065 A0A182M9E8 A0A182LFT6 A0A182FNH6 A0A0L0BSH4 B0X0Q1 A0A2K7P6A0 A0A182HB60 A0A1Q3FPP3 Q0IFL5 K7IZP3 A0A232FI12 T1J5A1 A0A1Y1L1P6 B4M578 W8CDU4 E9G328 E2AA51 Q5M8Z6 B4PSK4 A0A034VEK6 A0A0M5IZK8 E9IB02 A0A0A1X0R6 B3P2N5 B4QYD4 A0A0J7NMR8 B3M0W9 A0A0K8W8U8 A0A182IMR5 O97069 E2B6A2 A0A0P5IAU0 F7CD55 A0A0P5K2I4 Q7ZWM4 A0A452HV90 A0A1J1HXG2 A0A401SZR3 S4V5K7 A0A1W4V572 F6WVH7 A0A3Q2XDJ5 A0A2R5LK45 Q66I24 A0A293LYB3 A0A162P1R3 A0A1U7S2F0 G3SMU8 A0A341CVL9 A0A2Y9NJB3 A0A3Q2D7Y0 A0A131XZF6 A0A384B4K0 A0A2I0M414 A0A3Q7WYC8 A0A384CQ01 A0A088ACI2 G3PWQ7 A0A3Q4GPG8 U5YQ50 L8Y5S8 A0A060WCU7 G3PWR1 G1MCK8 B4JUJ3
D6X477 A0A336M4E0 A0A336MHX5 A0A2P8XPL1 A0A2J7QV22 A0A2J7QV24 U5EV09 J3JX12 A0A182SAU0 A0A182K946 A0A067QUJ2 W5J7W2 A0A182N639 A0A182Y4A8 A0A084VRC0 A0A2M4AI34 T1PCL4 A0A1B0CVH8 A0A182P5T4 A0A182RNR2 A0A1I8N6S5 A0A182Q4G0 A0A182VJI6 A0A182WRJ6 A0A182HU80 Q7PR38 A0A182TG76 A0A1I8PQ65 A0A182W065 A0A182M9E8 A0A182LFT6 A0A182FNH6 A0A0L0BSH4 B0X0Q1 A0A2K7P6A0 A0A182HB60 A0A1Q3FPP3 Q0IFL5 K7IZP3 A0A232FI12 T1J5A1 A0A1Y1L1P6 B4M578 W8CDU4 E9G328 E2AA51 Q5M8Z6 B4PSK4 A0A034VEK6 A0A0M5IZK8 E9IB02 A0A0A1X0R6 B3P2N5 B4QYD4 A0A0J7NMR8 B3M0W9 A0A0K8W8U8 A0A182IMR5 O97069 E2B6A2 A0A0P5IAU0 F7CD55 A0A0P5K2I4 Q7ZWM4 A0A452HV90 A0A1J1HXG2 A0A401SZR3 S4V5K7 A0A1W4V572 F6WVH7 A0A3Q2XDJ5 A0A2R5LK45 Q66I24 A0A293LYB3 A0A162P1R3 A0A1U7S2F0 G3SMU8 A0A341CVL9 A0A2Y9NJB3 A0A3Q2D7Y0 A0A131XZF6 A0A384B4K0 A0A2I0M414 A0A3Q7WYC8 A0A384CQ01 A0A088ACI2 G3PWQ7 A0A3Q4GPG8 U5YQ50 L8Y5S8 A0A060WCU7 G3PWR1 G1MCK8 B4JUJ3
EC Number
6.3.4.5
Pubmed
22118469
26354079
18362917
19820115
29403074
22516182
+ More
24845553 20920257 23761445 25244985 24438588 25315136 12364791 20966253 26108605 26483478 17510324 20075255 28648823 28004739 17994087 24495485 21292972 20798317 17550304 25348373 21282665 25830018 10731132 12537572 20431018 28562605 30297745 23371554 24813606 25186727 23385571 24755649 20010809
24845553 20920257 23761445 25244985 24438588 25315136 12364791 20966253 26108605 26483478 17510324 20075255 28648823 28004739 17994087 24495485 21292972 20798317 17550304 25348373 21282665 25830018 10731132 12537572 20431018 28562605 30297745 23371554 24813606 25186727 23385571 24755649 20010809
EMBL
NWSH01000105
PCG79517.1
ODYU01010139
SOQ55036.1
AGBW02009246
OWR51268.1
+ More
KQ461195 KPJ06692.1 RSAL01000268 RVE43214.1 KQ459596 KPI95501.1 KQ971410 EEZ97516.1 UFQT01000546 SSX25142.1 UFQT01001289 SSX29856.1 PYGN01001576 PSN33936.1 NEVH01010479 PNF32432.1 PNF32433.1 GANO01002084 JAB57787.1 BT127780 AEE62742.1 KK853400 KDR07860.1 ADMH02002109 ETN58945.1 ATLV01015601 KE525023 KFB40514.1 GGFK01007113 MBW40434.1 KA646429 AFP61058.1 AJWK01030592 AXCN02000193 APCN01000612 AAAB01008859 AXCM01000884 JRES01001422 KNC23030.1 DS232243 EDS38293.1 JXUM01031743 KQ560934 KXJ80285.1 GFDL01005464 JAV29581.1 CH477312 NNAY01000215 OXU29967.1 JH431855 GEZM01066974 JAV67609.1 CH940652 EDW59789.1 GAMC01001341 JAC05215.1 GL732530 EFX86402.1 GL438029 EFN69689.1 BC087767 CM000160 EDW96464.1 GAKP01018405 JAC40547.1 CP012526 ALC45768.1 GL762091 EFZ22280.1 GBXI01010064 JAD04228.1 CH954181 EDV48057.1 CM000364 EDX11891.1 LBMM01003212 KMQ93800.1 CH902617 EDV43198.2 GDHF01027485 GDHF01027252 GDHF01011616 GDHF01004855 JAI24829.1 JAI25062.1 JAI40698.1 JAI47459.1 AE001572 AE014297 BT060440 GL445930 EFN88795.1 GDIQ01220512 GDIQ01220511 GDIQ01220510 GDIQ01173620 GDIP01135803 JAK31215.1 JAL67911.1 AAMC01054407 AAMC01054408 AAMC01054409 AAMC01054410 AAMC01054411 AAMC01054412 AAMC01054413 GDIQ01242853 GDIQ01241406 GDIQ01238263 GDIQ01238262 GDIQ01207117 GDIQ01207116 GDIQ01207115 GDIQ01190570 GDIQ01190569 GDIQ01190568 GDIQ01099905 GDIQ01052873 GDIQ01033680 JAK61156.1 BC046941 CVRI01000020 CRK91214.1 BEZZ01000757 GCC35889.1 KC513735 AGO20698.1 GGLE01005760 MBY09886.1 BC081578 BC154282 AAH81578.1 GFWV01008933 MAA33662.1 LRGB01000512 KZS18447.1 GEFM01004276 JAP71520.1 AKCR02000043 PKK24425.1 KF444054 AGZ94902.2 KB364971 ELV11783.1 FR904427 CDQ63059.1 ACTA01001207 ACTA01177198 ACTA01185196 ACTA01193194 CH916374 EDV91163.1
KQ461195 KPJ06692.1 RSAL01000268 RVE43214.1 KQ459596 KPI95501.1 KQ971410 EEZ97516.1 UFQT01000546 SSX25142.1 UFQT01001289 SSX29856.1 PYGN01001576 PSN33936.1 NEVH01010479 PNF32432.1 PNF32433.1 GANO01002084 JAB57787.1 BT127780 AEE62742.1 KK853400 KDR07860.1 ADMH02002109 ETN58945.1 ATLV01015601 KE525023 KFB40514.1 GGFK01007113 MBW40434.1 KA646429 AFP61058.1 AJWK01030592 AXCN02000193 APCN01000612 AAAB01008859 AXCM01000884 JRES01001422 KNC23030.1 DS232243 EDS38293.1 JXUM01031743 KQ560934 KXJ80285.1 GFDL01005464 JAV29581.1 CH477312 NNAY01000215 OXU29967.1 JH431855 GEZM01066974 JAV67609.1 CH940652 EDW59789.1 GAMC01001341 JAC05215.1 GL732530 EFX86402.1 GL438029 EFN69689.1 BC087767 CM000160 EDW96464.1 GAKP01018405 JAC40547.1 CP012526 ALC45768.1 GL762091 EFZ22280.1 GBXI01010064 JAD04228.1 CH954181 EDV48057.1 CM000364 EDX11891.1 LBMM01003212 KMQ93800.1 CH902617 EDV43198.2 GDHF01027485 GDHF01027252 GDHF01011616 GDHF01004855 JAI24829.1 JAI25062.1 JAI40698.1 JAI47459.1 AE001572 AE014297 BT060440 GL445930 EFN88795.1 GDIQ01220512 GDIQ01220511 GDIQ01220510 GDIQ01173620 GDIP01135803 JAK31215.1 JAL67911.1 AAMC01054407 AAMC01054408 AAMC01054409 AAMC01054410 AAMC01054411 AAMC01054412 AAMC01054413 GDIQ01242853 GDIQ01241406 GDIQ01238263 GDIQ01238262 GDIQ01207117 GDIQ01207116 GDIQ01207115 GDIQ01190570 GDIQ01190569 GDIQ01190568 GDIQ01099905 GDIQ01052873 GDIQ01033680 JAK61156.1 BC046941 CVRI01000020 CRK91214.1 BEZZ01000757 GCC35889.1 KC513735 AGO20698.1 GGLE01005760 MBY09886.1 BC081578 BC154282 AAH81578.1 GFWV01008933 MAA33662.1 LRGB01000512 KZS18447.1 GEFM01004276 JAP71520.1 AKCR02000043 PKK24425.1 KF444054 AGZ94902.2 KB364971 ELV11783.1 FR904427 CDQ63059.1 ACTA01001207 ACTA01177198 ACTA01185196 ACTA01193194 CH916374 EDV91163.1
Proteomes
UP000218220
UP000007151
UP000053240
UP000283053
UP000053268
UP000007266
+ More
UP000245037 UP000235965 UP000075901 UP000075881 UP000027135 UP000000673 UP000075884 UP000076408 UP000030765 UP000092461 UP000075885 UP000075900 UP000095301 UP000075886 UP000075903 UP000076407 UP000075840 UP000007062 UP000075902 UP000095300 UP000075920 UP000075883 UP000075882 UP000069272 UP000037069 UP000002320 UP000069940 UP000249989 UP000008820 UP000002358 UP000215335 UP000008792 UP000000305 UP000000311 UP000008143 UP000002282 UP000092553 UP000008711 UP000000304 UP000036403 UP000007801 UP000075880 UP000000803 UP000008237 UP000291020 UP000183832 UP000287033 UP000192221 UP000264820 UP000000437 UP000076858 UP000189705 UP000007646 UP000252040 UP000248483 UP000265020 UP000261681 UP000053872 UP000286642 UP000261680 UP000291021 UP000005203 UP000007635 UP000261580 UP000011518 UP000193380 UP000008912 UP000001070
UP000245037 UP000235965 UP000075901 UP000075881 UP000027135 UP000000673 UP000075884 UP000076408 UP000030765 UP000092461 UP000075885 UP000075900 UP000095301 UP000075886 UP000075903 UP000076407 UP000075840 UP000007062 UP000075902 UP000095300 UP000075920 UP000075883 UP000075882 UP000069272 UP000037069 UP000002320 UP000069940 UP000249989 UP000008820 UP000002358 UP000215335 UP000008792 UP000000305 UP000000311 UP000008143 UP000002282 UP000092553 UP000008711 UP000000304 UP000036403 UP000007801 UP000075880 UP000000803 UP000008237 UP000291020 UP000183832 UP000287033 UP000192221 UP000264820 UP000000437 UP000076858 UP000189705 UP000007646 UP000252040 UP000248483 UP000265020 UP000261681 UP000053872 UP000286642 UP000261680 UP000291021 UP000005203 UP000007635 UP000261580 UP000011518 UP000193380 UP000008912 UP000001070
Pfam
PF00764 Arginosuc_synth
Interpro
Gene 3D
ProteinModelPortal
A0A2A4K5L2
A0A2H1WPY3
A0A212FC20
A0A194QMD4
A0A3S2NBM0
A0A194PPX9
+ More
D6X477 A0A336M4E0 A0A336MHX5 A0A2P8XPL1 A0A2J7QV22 A0A2J7QV24 U5EV09 J3JX12 A0A182SAU0 A0A182K946 A0A067QUJ2 W5J7W2 A0A182N639 A0A182Y4A8 A0A084VRC0 A0A2M4AI34 T1PCL4 A0A1B0CVH8 A0A182P5T4 A0A182RNR2 A0A1I8N6S5 A0A182Q4G0 A0A182VJI6 A0A182WRJ6 A0A182HU80 Q7PR38 A0A182TG76 A0A1I8PQ65 A0A182W065 A0A182M9E8 A0A182LFT6 A0A182FNH6 A0A0L0BSH4 B0X0Q1 A0A2K7P6A0 A0A182HB60 A0A1Q3FPP3 Q0IFL5 K7IZP3 A0A232FI12 T1J5A1 A0A1Y1L1P6 B4M578 W8CDU4 E9G328 E2AA51 Q5M8Z6 B4PSK4 A0A034VEK6 A0A0M5IZK8 E9IB02 A0A0A1X0R6 B3P2N5 B4QYD4 A0A0J7NMR8 B3M0W9 A0A0K8W8U8 A0A182IMR5 O97069 E2B6A2 A0A0P5IAU0 F7CD55 A0A0P5K2I4 Q7ZWM4 A0A452HV90 A0A1J1HXG2 A0A401SZR3 S4V5K7 A0A1W4V572 F6WVH7 A0A3Q2XDJ5 A0A2R5LK45 Q66I24 A0A293LYB3 A0A162P1R3 A0A1U7S2F0 G3SMU8 A0A341CVL9 A0A2Y9NJB3 A0A3Q2D7Y0 A0A131XZF6 A0A384B4K0 A0A2I0M414 A0A3Q7WYC8 A0A384CQ01 A0A088ACI2 G3PWQ7 A0A3Q4GPG8 U5YQ50 L8Y5S8 A0A060WCU7 G3PWR1 G1MCK8 B4JUJ3
D6X477 A0A336M4E0 A0A336MHX5 A0A2P8XPL1 A0A2J7QV22 A0A2J7QV24 U5EV09 J3JX12 A0A182SAU0 A0A182K946 A0A067QUJ2 W5J7W2 A0A182N639 A0A182Y4A8 A0A084VRC0 A0A2M4AI34 T1PCL4 A0A1B0CVH8 A0A182P5T4 A0A182RNR2 A0A1I8N6S5 A0A182Q4G0 A0A182VJI6 A0A182WRJ6 A0A182HU80 Q7PR38 A0A182TG76 A0A1I8PQ65 A0A182W065 A0A182M9E8 A0A182LFT6 A0A182FNH6 A0A0L0BSH4 B0X0Q1 A0A2K7P6A0 A0A182HB60 A0A1Q3FPP3 Q0IFL5 K7IZP3 A0A232FI12 T1J5A1 A0A1Y1L1P6 B4M578 W8CDU4 E9G328 E2AA51 Q5M8Z6 B4PSK4 A0A034VEK6 A0A0M5IZK8 E9IB02 A0A0A1X0R6 B3P2N5 B4QYD4 A0A0J7NMR8 B3M0W9 A0A0K8W8U8 A0A182IMR5 O97069 E2B6A2 A0A0P5IAU0 F7CD55 A0A0P5K2I4 Q7ZWM4 A0A452HV90 A0A1J1HXG2 A0A401SZR3 S4V5K7 A0A1W4V572 F6WVH7 A0A3Q2XDJ5 A0A2R5LK45 Q66I24 A0A293LYB3 A0A162P1R3 A0A1U7S2F0 G3SMU8 A0A341CVL9 A0A2Y9NJB3 A0A3Q2D7Y0 A0A131XZF6 A0A384B4K0 A0A2I0M414 A0A3Q7WYC8 A0A384CQ01 A0A088ACI2 G3PWQ7 A0A3Q4GPG8 U5YQ50 L8Y5S8 A0A060WCU7 G3PWR1 G1MCK8 B4JUJ3
PDB
2NZ2
E-value=1.83652e-134,
Score=1228
Ontologies
PATHWAY
GO
PANTHER
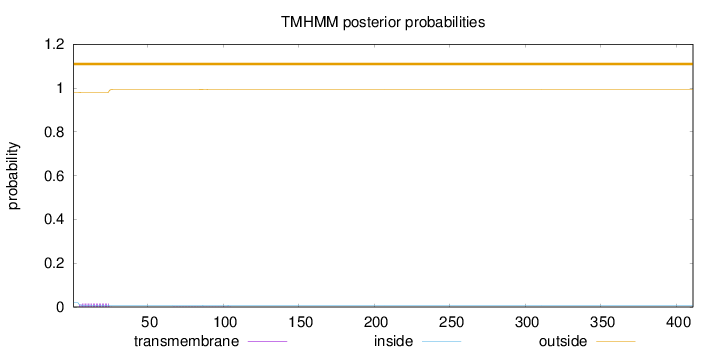
Topology
Subcellular location
Length:
411
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.32641
Exp number, first 60 AAs:
0.30359
Total prob of N-in:
0.01890
outside
1 - 411
Population Genetic Test Statistics
Pi
199.931688
Theta
148.831617
Tajima's D
1.030207
CLR
0
CSRT
0.665566721663917
Interpretation
Uncertain
