Gene
KWMTBOMO08786
Annotation
PREDICTED:_uncharacterized_protein_LOC106720812_[Papilio_machaon]
Location in the cell
Mitochondrial Reliability : 2.34
Sequence
CDS
ATGCAACGAACACAGGTCGAGAGCTTTTCTATAGAATTTCTTCATATCCTGAACTCGAAGCCTATTCCTCCGAACAGTCGTCTCAGAAAACTGTCACCGGCTTTGGGAGAGGACAAATTACTGCGGTTAGCCGGAAGAATTAAAGCCGCAGAAGGCATTGACCCTGACACGCGCTTCCCCGTCCTTCTTGACGGTCGTCACCCAATTGTACGTTTATTGGTACAATTTTATCATCGAAAAGCGGGTCACGCAAATTATGAACTAGTCGTGAACGAATTAAGACAAAAATTCTGGTTGTTAAGGCTGAGGAACACAGTGAGAACAGTAGCAAAAGAGTGTCCACTCTGCAGAATTAGAAAGTCCTTGCCGGTGAACCCGAAAACAGGAGATCTGCCACCTGAAAGATTAGCTCACCATCGACGGCCCTTTACTTTTACGGGTCTGGACTACTTCGGCCCCGTAAGCGCCACTATCGGGCGCAGGCATGAAAAACGTTATGTAGCCCTTTTCACCTGTTTAACTGTCAGAGCCCTTCACCTAGAAATGGTACATAGTCTATCGACTGACTCTGCTATCATGGCTATTAGAAGATTCATTGCAAGACGGGGCTGTCCAAACATTATTTTCTCGGACAACGGAACCTGCTTTGTTGGTGCCAATCGCATTCTCAGAGGTTTCTACGGAGAAAATGTCGAAGATTTCACAAGCACTCAAGGAATACTCTGGAAGTTCATCCCACCTGCGGCACCTAATTTCGGAGGATGTTGGGAGCGCTTGGTGCGATCAGTAAAGGTAGCTCTGAACGCAACATTAACCGAACGAGCTCCAAAAGAAGAAACCTTGAGCGTCTTACTAGCCGAGGCAGAAGCCATTGTCAATTCACGTCCATTAACTCATGTATCTACAGATCCTGACGACCCGACAACACTCACACCATTTCATTTCTTAATCGGTTCTTCGTCCGTGCAGACCTTACCTACTGCCTTGAACGATCACGATTTGGTCAGCCGTTCCGAATGGAGGAAAGCATTGAGGCTAGCAGATCATTTTTGGTCGCGTTGGATGCGCGAGGTTCTCCCGACCCTACAACCACGTGAGCAGTTAAGAGGCAACCGCAGTTCAACAGCTTTTCAAGAGGGTGACGTAGTAATTATTGCTGACTTTAACTTGCCCCGCGGAATATGGCCACGAGGACGTGTTGCGAAAGTGTACCCTGGGAAAGATGGCGTGATAAGGGTAGTGGACGTTGCAACAGCAGGGGGAATGCTACGCCGACCCGTACGAAAGCTGGTTAGGCTCTCCACCTAA
Protein
MQRTQVESFSIEFLHILNSKPIPPNSRLRKLSPALGEDKLLRLAGRIKAAEGIDPDTRFPVLLDGRHPIVRLLVQFYHRKAGHANYELVVNELRQKFWLLRLRNTVRTVAKECPLCRIRKSLPVNPKTGDLPPERLAHHRRPFTFTGLDYFGPVSATIGRRHEKRYVALFTCLTVRALHLEMVHSLSTDSAIMAIRRFIARRGCPNIIFSDNGTCFVGANRILRGFYGENVEDFTSTQGILWKFIPPAAPNFGGCWERLVRSVKVALNATLTERAPKEETLSVLLAEAEAIVNSRPLTHVSTDPDDPTTLTPFHFLIGSSSVQTLPTALNDHDLVSRSEWRKALRLADHFWSRWMREVLPTLQPREQLRGNRSSTAFQEGDVVIIADFNLPRGIWPRGRVAKVYPGKDGVIRVVDVATAGGMLRRPVRKLVRLST
Summary
Uniprot
A0A0N1IGH3
A0A3S2N5J7
Q9NDN0
Q9NDM9
A0A3S2P6R4
A0A2A4IRQ3
+ More
A0A2H1VMV3 Q2MGA5 A0A2W1BZX8 K7JA41 A0A226DGY5 A0A226D351 A0A226DH22 A0A182GD64 A0A146KYV8 A0A1Y1L683 W8AJC8 A0A226EFV7 A0A182HDR0 A0A182G4R3 A0A182GWN8 A0A182YLX0 A0A226E2P7 A0A226DM06 A0A182GRX9 A0A182GED3 A0A226EKF8 A0A182HER3 A0A182G9N0 A0A085LRU1 A0A1B0D7Y5 A0A182G2K7 A0A182HAK2 A0A1B0CTR4 A0A182G7Z0 A0A2M4BQP9 A0A085MRE0 A0A182GKP7 W4ZKP6 A0A1Y1LN76 A0A085NQS3 A0A182VT46 A0A2M4B8V2 A0A182RSB1 A0A1B0D864 A0A182XPX3 A0A085MU62 A0A182G5I1 A0A146TFB6 A0A182HAG0 A0A2M4B8T6 A0A182G6A0 A0A182N7R2 A0A0S7L6P4 A0A1B0DF44 A0A336MNL5 W4ZKP7 A0A336M9E8 A0A2M4CKZ0 A0A182G158 A0A2M4CKI9 A0A2M4BPT9 A0A034V9M7 A0A1S3SVP3 A0A085LTB9 A0A1W7R6F2 A0A453YZM1 A0A182XPK3 A0A182XPV1 A0A0N5E596 A0A182GY66 A0A182G1E0 A0A085NF60 A0A182GQC7 A0A182X1M1 A0A1S3K952 A0A085N4I6 I3KZ01 A0A2B4SX75 A0A085ND08 A0A182GPV7 A0A085M9S1 A0A085MSJ2 A0A182G9X5 W4ZDM7 A0A226EEI4 A0A336KMY3 A0A182H1N7 A0A085NCZ0 A0A182PWK8 A0A1W7R6D5 W4ZBS7 A0A0P6C5E3 A0A0P6AZF3 W4ZBS6 A0A182HCB4 A0A085N549 A0A085M9G8 A0A034WLY0 A0A146SEB9
A0A2H1VMV3 Q2MGA5 A0A2W1BZX8 K7JA41 A0A226DGY5 A0A226D351 A0A226DH22 A0A182GD64 A0A146KYV8 A0A1Y1L683 W8AJC8 A0A226EFV7 A0A182HDR0 A0A182G4R3 A0A182GWN8 A0A182YLX0 A0A226E2P7 A0A226DM06 A0A182GRX9 A0A182GED3 A0A226EKF8 A0A182HER3 A0A182G9N0 A0A085LRU1 A0A1B0D7Y5 A0A182G2K7 A0A182HAK2 A0A1B0CTR4 A0A182G7Z0 A0A2M4BQP9 A0A085MRE0 A0A182GKP7 W4ZKP6 A0A1Y1LN76 A0A085NQS3 A0A182VT46 A0A2M4B8V2 A0A182RSB1 A0A1B0D864 A0A182XPX3 A0A085MU62 A0A182G5I1 A0A146TFB6 A0A182HAG0 A0A2M4B8T6 A0A182G6A0 A0A182N7R2 A0A0S7L6P4 A0A1B0DF44 A0A336MNL5 W4ZKP7 A0A336M9E8 A0A2M4CKZ0 A0A182G158 A0A2M4CKI9 A0A2M4BPT9 A0A034V9M7 A0A1S3SVP3 A0A085LTB9 A0A1W7R6F2 A0A453YZM1 A0A182XPK3 A0A182XPV1 A0A0N5E596 A0A182GY66 A0A182G1E0 A0A085NF60 A0A182GQC7 A0A182X1M1 A0A1S3K952 A0A085N4I6 I3KZ01 A0A2B4SX75 A0A085ND08 A0A182GPV7 A0A085M9S1 A0A085MSJ2 A0A182G9X5 W4ZDM7 A0A226EEI4 A0A336KMY3 A0A182H1N7 A0A085NCZ0 A0A182PWK8 A0A1W7R6D5 W4ZBS7 A0A0P6C5E3 A0A0P6AZF3 W4ZBS6 A0A182HCB4 A0A085N549 A0A085M9G8 A0A034WLY0 A0A146SEB9
Pubmed
EMBL
KQ459986
KPJ18861.1
RSAL01000375
RVE42106.1
AB042118
BAA95569.1
+ More
AB042119 BAA95570.1 RSAL01000010 RVE53755.1 NWSH01008670 PCG62455.1 ODYU01003174 SOQ41584.1 AF530470 AAQ09229.1 KZ149895 PZC78807.1 AAZX01023278 LNIX01000019 OXA44812.1 LNIX01000039 OXA39294.1 OXA44855.1 JXUM01009840 KQ560293 KXJ83223.1 GDHC01018323 JAQ00306.1 GEZM01063488 GEZM01063487 JAV69084.1 GAMC01017930 JAB88625.1 LNIX01000004 OXA56250.1 JXUM01130316 KQ567562 KXJ69370.1 JXUM01144298 KQ569759 KXJ68523.1 JXUM01020113 KQ560564 KXJ81881.1 LNIX01000007 OXA52015.1 LNIX01000015 OXA46575.1 JXUM01083542 KQ563383 KXJ73941.1 JXUM01162450 KQ574455 KXJ67869.1 LNIX01000003 OXA57597.1 JXUM01005801 KQ560202 KXJ83866.1 JXUM01049477 KQ561606 KXJ78017.1 KL363318 KFD47687.1 AJVK01033431 JXUM01139649 KQ568932 KXJ68836.1 JXUM01122819 KQ566647 KXJ69950.1 AJWK01027898 JXUM01151198 KQ571164 KXJ68235.1 GGFJ01005957 MBW55098.1 KL367737 KFD59786.1 JXUM01013377 KQ560375 KXJ82728.1 AAGJ04095911 GEZM01052586 JAV74401.1 KL367480 KFD71819.1 GGFJ01000333 MBW49474.1 AJVK01037041 KL367650 KFD60758.1 JXUM01146045 JXUM01146046 GCES01094753 JAQ91569.1 JXUM01031097 KQ560909 KXJ80382.1 GGFJ01000306 MBW49447.1 JXUM01147607 KQ570400 KXJ68371.1 GBYX01183181 JAO85198.1 AJVK01058515 UFQS01001404 UFQT01001404 SSX10721.1 SSX30403.1 UFQT01000590 SSX25539.1 GGFL01001842 MBW66020.1 JXUM01136032 KQ568367 KXJ69026.1 GGFL01001682 MBW65860.1 GGFJ01005956 MBW55097.1 GAKP01020709 JAC38243.1 KL363300 KFD48215.1 GEHC01000913 JAV46732.1 AAAB01008847 JXUM01020939 KQ560586 KXJ81781.1 JXUM01038372 KQ561162 KXJ79503.1 KL367508 KFD68106.1 JXUM01080154 KQ563168 KXJ74353.1 KL367556 KFD64382.1 AERX01020956 LSMT01000016 PFX33015.1 KL367515 KFD67354.1 JXUM01016534 JXUM01056823 KQ561926 KQ560461 KXJ77135.1 KXJ82302.1 KL363212 KFD53967.1 KL367687 KFD60188.1 JXUM01049933 KQ561627 KXJ77955.1 AAGJ04011066 OXA55241.1 UFQS01000284 UFQT01000284 SSX02410.1 SSX22785.1 JXUM01103818 KQ564835 KXJ71674.1 KFD67336.1 GEHC01000921 JAV46724.1 AAGJ04122915 GDIP01004568 JAM99147.1 GDIP01036233 JAM67482.1 JXUM01127084 KQ567147 KXJ69602.1 KL367553 KFD64595.1 KFD53864.1 GAKP01004189 JAC54763.1 GCES01107353 JAQ78969.1
AB042119 BAA95570.1 RSAL01000010 RVE53755.1 NWSH01008670 PCG62455.1 ODYU01003174 SOQ41584.1 AF530470 AAQ09229.1 KZ149895 PZC78807.1 AAZX01023278 LNIX01000019 OXA44812.1 LNIX01000039 OXA39294.1 OXA44855.1 JXUM01009840 KQ560293 KXJ83223.1 GDHC01018323 JAQ00306.1 GEZM01063488 GEZM01063487 JAV69084.1 GAMC01017930 JAB88625.1 LNIX01000004 OXA56250.1 JXUM01130316 KQ567562 KXJ69370.1 JXUM01144298 KQ569759 KXJ68523.1 JXUM01020113 KQ560564 KXJ81881.1 LNIX01000007 OXA52015.1 LNIX01000015 OXA46575.1 JXUM01083542 KQ563383 KXJ73941.1 JXUM01162450 KQ574455 KXJ67869.1 LNIX01000003 OXA57597.1 JXUM01005801 KQ560202 KXJ83866.1 JXUM01049477 KQ561606 KXJ78017.1 KL363318 KFD47687.1 AJVK01033431 JXUM01139649 KQ568932 KXJ68836.1 JXUM01122819 KQ566647 KXJ69950.1 AJWK01027898 JXUM01151198 KQ571164 KXJ68235.1 GGFJ01005957 MBW55098.1 KL367737 KFD59786.1 JXUM01013377 KQ560375 KXJ82728.1 AAGJ04095911 GEZM01052586 JAV74401.1 KL367480 KFD71819.1 GGFJ01000333 MBW49474.1 AJVK01037041 KL367650 KFD60758.1 JXUM01146045 JXUM01146046 GCES01094753 JAQ91569.1 JXUM01031097 KQ560909 KXJ80382.1 GGFJ01000306 MBW49447.1 JXUM01147607 KQ570400 KXJ68371.1 GBYX01183181 JAO85198.1 AJVK01058515 UFQS01001404 UFQT01001404 SSX10721.1 SSX30403.1 UFQT01000590 SSX25539.1 GGFL01001842 MBW66020.1 JXUM01136032 KQ568367 KXJ69026.1 GGFL01001682 MBW65860.1 GGFJ01005956 MBW55097.1 GAKP01020709 JAC38243.1 KL363300 KFD48215.1 GEHC01000913 JAV46732.1 AAAB01008847 JXUM01020939 KQ560586 KXJ81781.1 JXUM01038372 KQ561162 KXJ79503.1 KL367508 KFD68106.1 JXUM01080154 KQ563168 KXJ74353.1 KL367556 KFD64382.1 AERX01020956 LSMT01000016 PFX33015.1 KL367515 KFD67354.1 JXUM01016534 JXUM01056823 KQ561926 KQ560461 KXJ77135.1 KXJ82302.1 KL363212 KFD53967.1 KL367687 KFD60188.1 JXUM01049933 KQ561627 KXJ77955.1 AAGJ04011066 OXA55241.1 UFQS01000284 UFQT01000284 SSX02410.1 SSX22785.1 JXUM01103818 KQ564835 KXJ71674.1 KFD67336.1 GEHC01000921 JAV46724.1 AAGJ04122915 GDIP01004568 JAM99147.1 GDIP01036233 JAM67482.1 JXUM01127084 KQ567147 KXJ69602.1 KL367553 KFD64595.1 KFD53864.1 GAKP01004189 JAC54763.1 GCES01107353 JAQ78969.1
Proteomes
Pfam
Interpro
IPR036397
RNaseH_sf
+ More
IPR041588 Integrase_H2C2
IPR040676 DUF5641
IPR008737 Peptidase_asp_put
IPR008042 Retrotrans_Pao
IPR012337 RNaseH-like_sf
IPR001995 Peptidase_A2_cat
IPR001584 Integrase_cat-core
IPR005312 DUF1759
IPR036305 RGS_sf
IPR000961 AGC-kinase_C
IPR000719 Prot_kinase_dom
IPR000239 GPCR_kinase
IPR017441 Protein_kinase_ATP_BS
IPR011009 Kinase-like_dom_sf
IPR016137 RGS
IPR001878 Znf_CCHC
IPR006094 Oxid_FAD_bind_N
IPR016166 FAD-bd_PCMH
IPR016164 FAD-linked_Oxase-like_C
IPR016170 Cytok_DH_C_sf
IPR015213 Cholesterol_OX_subst-bd
IPR036318 FAD-bd_PCMH-like_sf
IPR016169 FAD-bd_PCMH_sub2
IPR016167 FAD-bd_PCMH_sub1
IPR019787 Znf_PHD-finger
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR011011 Znf_FYVE_PHD
IPR009563 SSSCA1
IPR000477 RT_dom
IPR019786 Zinc_finger_PHD-type_CS
IPR006594 LisH
IPR006595 CTLH_C
IPR024964 CTLH/CRA
IPR021109 Peptidase_aspartic_dom_sf
IPR041588 Integrase_H2C2
IPR040676 DUF5641
IPR008737 Peptidase_asp_put
IPR008042 Retrotrans_Pao
IPR012337 RNaseH-like_sf
IPR001995 Peptidase_A2_cat
IPR001584 Integrase_cat-core
IPR005312 DUF1759
IPR036305 RGS_sf
IPR000961 AGC-kinase_C
IPR000719 Prot_kinase_dom
IPR000239 GPCR_kinase
IPR017441 Protein_kinase_ATP_BS
IPR011009 Kinase-like_dom_sf
IPR016137 RGS
IPR001878 Znf_CCHC
IPR006094 Oxid_FAD_bind_N
IPR016166 FAD-bd_PCMH
IPR016164 FAD-linked_Oxase-like_C
IPR016170 Cytok_DH_C_sf
IPR015213 Cholesterol_OX_subst-bd
IPR036318 FAD-bd_PCMH-like_sf
IPR016169 FAD-bd_PCMH_sub2
IPR016167 FAD-bd_PCMH_sub1
IPR019787 Znf_PHD-finger
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR011011 Znf_FYVE_PHD
IPR009563 SSSCA1
IPR000477 RT_dom
IPR019786 Zinc_finger_PHD-type_CS
IPR006594 LisH
IPR006595 CTLH_C
IPR024964 CTLH/CRA
IPR021109 Peptidase_aspartic_dom_sf
SUPFAM
ProteinModelPortal
A0A0N1IGH3
A0A3S2N5J7
Q9NDN0
Q9NDM9
A0A3S2P6R4
A0A2A4IRQ3
+ More
A0A2H1VMV3 Q2MGA5 A0A2W1BZX8 K7JA41 A0A226DGY5 A0A226D351 A0A226DH22 A0A182GD64 A0A146KYV8 A0A1Y1L683 W8AJC8 A0A226EFV7 A0A182HDR0 A0A182G4R3 A0A182GWN8 A0A182YLX0 A0A226E2P7 A0A226DM06 A0A182GRX9 A0A182GED3 A0A226EKF8 A0A182HER3 A0A182G9N0 A0A085LRU1 A0A1B0D7Y5 A0A182G2K7 A0A182HAK2 A0A1B0CTR4 A0A182G7Z0 A0A2M4BQP9 A0A085MRE0 A0A182GKP7 W4ZKP6 A0A1Y1LN76 A0A085NQS3 A0A182VT46 A0A2M4B8V2 A0A182RSB1 A0A1B0D864 A0A182XPX3 A0A085MU62 A0A182G5I1 A0A146TFB6 A0A182HAG0 A0A2M4B8T6 A0A182G6A0 A0A182N7R2 A0A0S7L6P4 A0A1B0DF44 A0A336MNL5 W4ZKP7 A0A336M9E8 A0A2M4CKZ0 A0A182G158 A0A2M4CKI9 A0A2M4BPT9 A0A034V9M7 A0A1S3SVP3 A0A085LTB9 A0A1W7R6F2 A0A453YZM1 A0A182XPK3 A0A182XPV1 A0A0N5E596 A0A182GY66 A0A182G1E0 A0A085NF60 A0A182GQC7 A0A182X1M1 A0A1S3K952 A0A085N4I6 I3KZ01 A0A2B4SX75 A0A085ND08 A0A182GPV7 A0A085M9S1 A0A085MSJ2 A0A182G9X5 W4ZDM7 A0A226EEI4 A0A336KMY3 A0A182H1N7 A0A085NCZ0 A0A182PWK8 A0A1W7R6D5 W4ZBS7 A0A0P6C5E3 A0A0P6AZF3 W4ZBS6 A0A182HCB4 A0A085N549 A0A085M9G8 A0A034WLY0 A0A146SEB9
A0A2H1VMV3 Q2MGA5 A0A2W1BZX8 K7JA41 A0A226DGY5 A0A226D351 A0A226DH22 A0A182GD64 A0A146KYV8 A0A1Y1L683 W8AJC8 A0A226EFV7 A0A182HDR0 A0A182G4R3 A0A182GWN8 A0A182YLX0 A0A226E2P7 A0A226DM06 A0A182GRX9 A0A182GED3 A0A226EKF8 A0A182HER3 A0A182G9N0 A0A085LRU1 A0A1B0D7Y5 A0A182G2K7 A0A182HAK2 A0A1B0CTR4 A0A182G7Z0 A0A2M4BQP9 A0A085MRE0 A0A182GKP7 W4ZKP6 A0A1Y1LN76 A0A085NQS3 A0A182VT46 A0A2M4B8V2 A0A182RSB1 A0A1B0D864 A0A182XPX3 A0A085MU62 A0A182G5I1 A0A146TFB6 A0A182HAG0 A0A2M4B8T6 A0A182G6A0 A0A182N7R2 A0A0S7L6P4 A0A1B0DF44 A0A336MNL5 W4ZKP7 A0A336M9E8 A0A2M4CKZ0 A0A182G158 A0A2M4CKI9 A0A2M4BPT9 A0A034V9M7 A0A1S3SVP3 A0A085LTB9 A0A1W7R6F2 A0A453YZM1 A0A182XPK3 A0A182XPV1 A0A0N5E596 A0A182GY66 A0A182G1E0 A0A085NF60 A0A182GQC7 A0A182X1M1 A0A1S3K952 A0A085N4I6 I3KZ01 A0A2B4SX75 A0A085ND08 A0A182GPV7 A0A085M9S1 A0A085MSJ2 A0A182G9X5 W4ZDM7 A0A226EEI4 A0A336KMY3 A0A182H1N7 A0A085NCZ0 A0A182PWK8 A0A1W7R6D5 W4ZBS7 A0A0P6C5E3 A0A0P6AZF3 W4ZBS6 A0A182HCB4 A0A085N549 A0A085M9G8 A0A034WLY0 A0A146SEB9
Ontologies
KEGG
GO
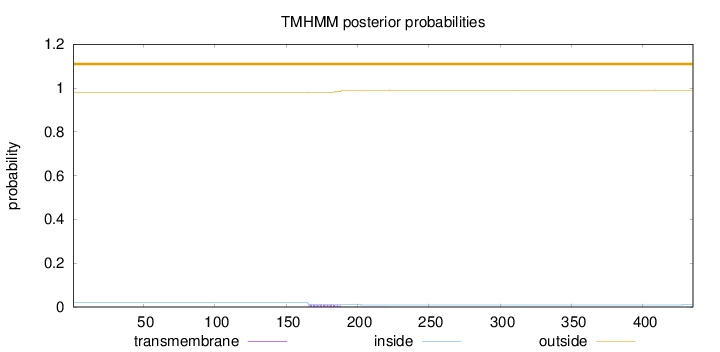
Topology
Length:
435
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.23009
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01995
outside
1 - 435
Population Genetic Test Statistics
Pi
226.340108
Theta
170.410555
Tajima's D
1.094967
CLR
0.503198
CSRT
0.687065646717664
Interpretation
Uncertain
