Gene
KWMTBOMO08784
Annotation
endonuclease-reverse_transcriptase_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 2.144
Sequence
CDS
ATGAAATTAATGGTGGTGGATCGTGCAAATGTTCTAAGATTGAATGGGGCGCTTGGATTAGAGGTTGTAGACGAATTCGTGTATCTCGGGTCCTGCATCAGCAACAAAGGCTCATGTGAAACTGACGTGCGCCGACGTATCGGAATGGCTAAAAGTGCGATGTCTCAGCTACAGAAGATCTGGAAGAATAGAGGGATAGGCCTGAAAACTAAAACCAGACTGGTGCGGACTCTTGTTTTCTCTATCTTCTTGTATGGAGCTGAGACATGGACGTTGAAAGCAGCAGACCGAATGCGCATCGATGCTTTCGAAATGTGGTGTTGGAGACGCATGCTAAGAATTCCGTGGACAGCCTTCCGGACTAACGTGTCGATATTGAAGCAACTGGGCATCAAAATCAGACTGTCTACTGTATGTCTGAAGCGGATCCTCGAATATTTTGGTCACATTGCGAGGAAAGACGGCGATAACCTGGAAAAACTTATGATCACAGGGAAAGTTGAAGGCAACAGATCTAGAGGCCGCAACCATACTCGTTGGTCCGATCAGATCCGCACCGCTTTGGACAGTACAGTGTACAGCGCGCTGCATGACGCCGTGGACAGAGGGAAAGTGGAAAAAGATCCTAAGATTGAAGCTCATGGGTGA
Protein
MKLMVVDRANVLRLNGALGLEVVDEFVYLGSCISNKGSCETDVRRRIGMAKSAMSQLQKIWKNRGIGLKTKTRLVRTLVFSIFLYGAETWTLKAADRMRIDAFEMWCWRRMLRIPWTAFRTNVSILKQLGIKIRLSTVCLKRILEYFGHIARKDGDNLEKLMITGKVEGNRSRGRNHTRWSDQIRTALDSTVYSALHDAVDRGKVEKDPKIEAHG
Summary
Uniprot
D7F163
D7F172
A0A2H1VU13
D7F168
D7F161
D7F162
+ More
A0A3S2L0R7 A0A2H1V742 A0A2W1BGD9 D5LB39 D7F175 A0A2H1VNK5 D7F167 A0A437AU38 A0A2G8JE23 A0A2G8JVI4 A0A402EVI9 A0A2G8JVA1 A0A2G8K183 A0A2G8L6P4 A0A2G8K796 A0A402EPS0 A0A402ELI2 A0A402F820 A0A402EUI3 A0A402EKM6 A0A402F7K5 A0A402EU36 A0A402EU18 A0A402F4B4 A0A402EZ91 A0A402EVB7 A0A402EX62 A0A402F218 A0A402FFT5 A0A402EL74 A0A402EKQ9 A0A402F0Z3 A0A402FRA5 A0A402E7Y2 A0A402EP91 A0A402EZQ5 A0A402EBN8 A0A402EL02 A0A402F9C0 A0A402EJN5 A0A402F3R2 A0A402F0P9 A0A402EJ80 A0A402EBI1 A0A402FA63 A0A402EKH9 A0A402F360 A0A402F6A8 A0A402EWU1 A0A402EZ24 A0A402EKQ0 A0A402EKS1 A0A402F524 A0A402EKM7 A0A402F166 A0A402FL85 A0A402ECE7 A0A402FDS4 A0A402FJR0 A0A402FWJ9 A0A402EZT0 A0A402ETF8 A0A402EH06 A0A402F0A9 A0A402EJ24 A0A402ECI4 A0A402FDL1 A0A402EBU4 A0A402F4P0 A0A402E850 A0A402EL56 A0A402FLC1 A0A402EBM0 A0A402ET00 A0A402F1V1 A0A402EYN6 A0A402FLP7 A0A402EQH3 A0A402EBL6 A0A402F241 A0A402EZ17 A0A402EKD3 A0A402E7Y8 A0A402EWI9 A0A402ECT4 A0A402EWP4 A0A402EZI8 A0A402FN33 A0A402EL71 A0A402EW85 A0A402FFD1 A0A402EBA1 A0A402FRC2 A0A402E6L1
A0A3S2L0R7 A0A2H1V742 A0A2W1BGD9 D5LB39 D7F175 A0A2H1VNK5 D7F167 A0A437AU38 A0A2G8JE23 A0A2G8JVI4 A0A402EVI9 A0A2G8JVA1 A0A2G8K183 A0A2G8L6P4 A0A2G8K796 A0A402EPS0 A0A402ELI2 A0A402F820 A0A402EUI3 A0A402EKM6 A0A402F7K5 A0A402EU36 A0A402EU18 A0A402F4B4 A0A402EZ91 A0A402EVB7 A0A402EX62 A0A402F218 A0A402FFT5 A0A402EL74 A0A402EKQ9 A0A402F0Z3 A0A402FRA5 A0A402E7Y2 A0A402EP91 A0A402EZQ5 A0A402EBN8 A0A402EL02 A0A402F9C0 A0A402EJN5 A0A402F3R2 A0A402F0P9 A0A402EJ80 A0A402EBI1 A0A402FA63 A0A402EKH9 A0A402F360 A0A402F6A8 A0A402EWU1 A0A402EZ24 A0A402EKQ0 A0A402EKS1 A0A402F524 A0A402EKM7 A0A402F166 A0A402FL85 A0A402ECE7 A0A402FDS4 A0A402FJR0 A0A402FWJ9 A0A402EZT0 A0A402ETF8 A0A402EH06 A0A402F0A9 A0A402EJ24 A0A402ECI4 A0A402FDL1 A0A402EBU4 A0A402F4P0 A0A402E850 A0A402EL56 A0A402FLC1 A0A402EBM0 A0A402ET00 A0A402F1V1 A0A402EYN6 A0A402FLP7 A0A402EQH3 A0A402EBL6 A0A402F241 A0A402EZ17 A0A402EKD3 A0A402E7Y8 A0A402EWI9 A0A402ECT4 A0A402EWP4 A0A402EZI8 A0A402FN33 A0A402EL71 A0A402EW85 A0A402FFD1 A0A402EBA1 A0A402FRC2 A0A402E6L1
EMBL
FJ265548
ADI61816.1
FJ265557
ADI61825.1
ODYU01004401
SOQ44236.1
+ More
FJ265553 ADI61821.1 FJ265546 ADI61814.1 FJ265547 ADI61815.1 RSAL01000289 RVE42931.1 ODYU01001007 SOQ36619.1 KZ150203 PZC72267.1 GU815090 ADF18553.1 FJ265560 ADI61828.1 ODYU01003523 SOQ42419.1 FJ265552 ADI61820.1 RSAL01000427 RVE41762.1 MRZV01002322 PIK33998.1 MRZV01001207 PIK39730.1 BDOT01000049 GCF48199.1 MRZV01001216 PIK39650.1 MRZV01000994 PIK41715.1 MRZV01000196 PIK55924.1 MRZV01000817 PIK43880.1 BDOT01000029 GCF46213.1 BDOT01000022 GCF45060.1 BDOT01000136 GCF52589.1 BDOT01000045 GCF47862.1 BDOT01000018 GCF44776.1 BDOT01000127 GCF52250.1 BDOT01000043 GCF47709.1 GCF47683.1 BDOT01000102 GCF51323.1 BDOT01000067 GCF49499.1 GCF48161.1 BDOT01000058 GCF48816.1 BDOT01000084 GCF50558.1 BDOT01000240 GCF55388.1 BDOT01000020 GCF44950.1 GCF44774.1 BDOT01000075 GCF50047.1 BDOT01000476 GCF58985.1 BDOT01000002 GCF40281.1 BDOT01000028 GCF46025.1 BDOT01000069 GCF49671.1 BDOT01000005 GCF41659.1 GCF44896.1 BDOT01000150 GCF53026.1 BDOT01000016 BDOT01001142 GCF44381.1 GCF61716.1 BDOT01000096 GCF51106.1 BDOT01000074 GCF50011.1 BDOT01000015 GCF44244.1 GCF41533.1 BDOT01000161 GCF53338.1 GCF44715.1 BDOT01000090 GCF50896.1 BDOT01000123 GCF52043.1 BDOT01000055 GCF48663.1 BDOT01000065 GCF49384.1 GCF44764.1 GCF44759.1 BDOT01000110 GCF47846.1 GCF51642.1 GCF44750.1 BDOT01000078 GCF50245.1 BDOT01000333 GCF57251.1 BDOT01000006 GCF41929.1 BDOT01000024 BDOT01000208 GCF45062.1 GCF45289.1 GCF54601.1 BDOT01000298 GCF56713.1 BDOT01000816 GCF60864.1 GCF49634.1 BDOT01000040 GCF47495.1 BDOT01000011 GCF43472.1 BDOT01000073 GCF49926.1 GCF44200.1 GCF41907.1 BDOT01000203 GCF54495.1 GCF41638.1 GCF49423.1 BDOT01000105 GCF51453.1 GCF40389.1 GCF44946.1 BDOT01000335 GCF57283.1 GCF41639.1 BDOT01000038 GCF47337.1 BDOT01000082 GCF50401.1 BDOT01000063 GCF49303.1 GCF57285.1 BDOT01000031 GCF46419.1 GCF41567.1 BDOT01000083 GCF50491.1 BDOT01000066 GCF49478.1 BDOT01000017 GCF44644.1 GCF40329.1 BDOT01000054 GCF48586.1 GCF41954.1 GCF48642.1 GCF49500.1 BDOT01000374 GCF57846.1 GCF44937.1 BDOT01000052 GCF48459.1 BDOT01000234 GCF55219.1 GCF41453.1 BDOT01000484 GCF59029.1 BDOT01000001 GCF39783.1
FJ265553 ADI61821.1 FJ265546 ADI61814.1 FJ265547 ADI61815.1 RSAL01000289 RVE42931.1 ODYU01001007 SOQ36619.1 KZ150203 PZC72267.1 GU815090 ADF18553.1 FJ265560 ADI61828.1 ODYU01003523 SOQ42419.1 FJ265552 ADI61820.1 RSAL01000427 RVE41762.1 MRZV01002322 PIK33998.1 MRZV01001207 PIK39730.1 BDOT01000049 GCF48199.1 MRZV01001216 PIK39650.1 MRZV01000994 PIK41715.1 MRZV01000196 PIK55924.1 MRZV01000817 PIK43880.1 BDOT01000029 GCF46213.1 BDOT01000022 GCF45060.1 BDOT01000136 GCF52589.1 BDOT01000045 GCF47862.1 BDOT01000018 GCF44776.1 BDOT01000127 GCF52250.1 BDOT01000043 GCF47709.1 GCF47683.1 BDOT01000102 GCF51323.1 BDOT01000067 GCF49499.1 GCF48161.1 BDOT01000058 GCF48816.1 BDOT01000084 GCF50558.1 BDOT01000240 GCF55388.1 BDOT01000020 GCF44950.1 GCF44774.1 BDOT01000075 GCF50047.1 BDOT01000476 GCF58985.1 BDOT01000002 GCF40281.1 BDOT01000028 GCF46025.1 BDOT01000069 GCF49671.1 BDOT01000005 GCF41659.1 GCF44896.1 BDOT01000150 GCF53026.1 BDOT01000016 BDOT01001142 GCF44381.1 GCF61716.1 BDOT01000096 GCF51106.1 BDOT01000074 GCF50011.1 BDOT01000015 GCF44244.1 GCF41533.1 BDOT01000161 GCF53338.1 GCF44715.1 BDOT01000090 GCF50896.1 BDOT01000123 GCF52043.1 BDOT01000055 GCF48663.1 BDOT01000065 GCF49384.1 GCF44764.1 GCF44759.1 BDOT01000110 GCF47846.1 GCF51642.1 GCF44750.1 BDOT01000078 GCF50245.1 BDOT01000333 GCF57251.1 BDOT01000006 GCF41929.1 BDOT01000024 BDOT01000208 GCF45062.1 GCF45289.1 GCF54601.1 BDOT01000298 GCF56713.1 BDOT01000816 GCF60864.1 GCF49634.1 BDOT01000040 GCF47495.1 BDOT01000011 GCF43472.1 BDOT01000073 GCF49926.1 GCF44200.1 GCF41907.1 BDOT01000203 GCF54495.1 GCF41638.1 GCF49423.1 BDOT01000105 GCF51453.1 GCF40389.1 GCF44946.1 BDOT01000335 GCF57283.1 GCF41639.1 BDOT01000038 GCF47337.1 BDOT01000082 GCF50401.1 BDOT01000063 GCF49303.1 GCF57285.1 BDOT01000031 GCF46419.1 GCF41567.1 BDOT01000083 GCF50491.1 BDOT01000066 GCF49478.1 BDOT01000017 GCF44644.1 GCF40329.1 BDOT01000054 GCF48586.1 GCF41954.1 GCF48642.1 GCF49500.1 BDOT01000374 GCF57846.1 GCF44937.1 BDOT01000052 GCF48459.1 BDOT01000234 GCF55219.1 GCF41453.1 BDOT01000484 GCF59029.1 BDOT01000001 GCF39783.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
D7F163
D7F172
A0A2H1VU13
D7F168
D7F161
D7F162
+ More
A0A3S2L0R7 A0A2H1V742 A0A2W1BGD9 D5LB39 D7F175 A0A2H1VNK5 D7F167 A0A437AU38 A0A2G8JE23 A0A2G8JVI4 A0A402EVI9 A0A2G8JVA1 A0A2G8K183 A0A2G8L6P4 A0A2G8K796 A0A402EPS0 A0A402ELI2 A0A402F820 A0A402EUI3 A0A402EKM6 A0A402F7K5 A0A402EU36 A0A402EU18 A0A402F4B4 A0A402EZ91 A0A402EVB7 A0A402EX62 A0A402F218 A0A402FFT5 A0A402EL74 A0A402EKQ9 A0A402F0Z3 A0A402FRA5 A0A402E7Y2 A0A402EP91 A0A402EZQ5 A0A402EBN8 A0A402EL02 A0A402F9C0 A0A402EJN5 A0A402F3R2 A0A402F0P9 A0A402EJ80 A0A402EBI1 A0A402FA63 A0A402EKH9 A0A402F360 A0A402F6A8 A0A402EWU1 A0A402EZ24 A0A402EKQ0 A0A402EKS1 A0A402F524 A0A402EKM7 A0A402F166 A0A402FL85 A0A402ECE7 A0A402FDS4 A0A402FJR0 A0A402FWJ9 A0A402EZT0 A0A402ETF8 A0A402EH06 A0A402F0A9 A0A402EJ24 A0A402ECI4 A0A402FDL1 A0A402EBU4 A0A402F4P0 A0A402E850 A0A402EL56 A0A402FLC1 A0A402EBM0 A0A402ET00 A0A402F1V1 A0A402EYN6 A0A402FLP7 A0A402EQH3 A0A402EBL6 A0A402F241 A0A402EZ17 A0A402EKD3 A0A402E7Y8 A0A402EWI9 A0A402ECT4 A0A402EWP4 A0A402EZI8 A0A402FN33 A0A402EL71 A0A402EW85 A0A402FFD1 A0A402EBA1 A0A402FRC2 A0A402E6L1
A0A3S2L0R7 A0A2H1V742 A0A2W1BGD9 D5LB39 D7F175 A0A2H1VNK5 D7F167 A0A437AU38 A0A2G8JE23 A0A2G8JVI4 A0A402EVI9 A0A2G8JVA1 A0A2G8K183 A0A2G8L6P4 A0A2G8K796 A0A402EPS0 A0A402ELI2 A0A402F820 A0A402EUI3 A0A402EKM6 A0A402F7K5 A0A402EU36 A0A402EU18 A0A402F4B4 A0A402EZ91 A0A402EVB7 A0A402EX62 A0A402F218 A0A402FFT5 A0A402EL74 A0A402EKQ9 A0A402F0Z3 A0A402FRA5 A0A402E7Y2 A0A402EP91 A0A402EZQ5 A0A402EBN8 A0A402EL02 A0A402F9C0 A0A402EJN5 A0A402F3R2 A0A402F0P9 A0A402EJ80 A0A402EBI1 A0A402FA63 A0A402EKH9 A0A402F360 A0A402F6A8 A0A402EWU1 A0A402EZ24 A0A402EKQ0 A0A402EKS1 A0A402F524 A0A402EKM7 A0A402F166 A0A402FL85 A0A402ECE7 A0A402FDS4 A0A402FJR0 A0A402FWJ9 A0A402EZT0 A0A402ETF8 A0A402EH06 A0A402F0A9 A0A402EJ24 A0A402ECI4 A0A402FDL1 A0A402EBU4 A0A402F4P0 A0A402E850 A0A402EL56 A0A402FLC1 A0A402EBM0 A0A402ET00 A0A402F1V1 A0A402EYN6 A0A402FLP7 A0A402EQH3 A0A402EBL6 A0A402F241 A0A402EZ17 A0A402EKD3 A0A402E7Y8 A0A402EWI9 A0A402ECT4 A0A402EWP4 A0A402EZI8 A0A402FN33 A0A402EL71 A0A402EW85 A0A402FFD1 A0A402EBA1 A0A402FRC2 A0A402E6L1
Ontologies
PANTHER
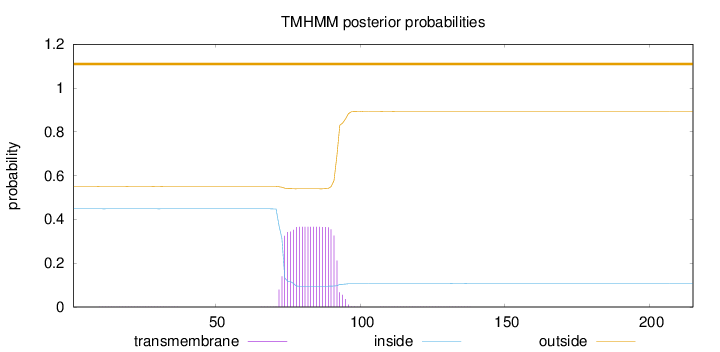
Topology
Length:
215
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
7.07512
Exp number, first 60 AAs:
0.01651
Total prob of N-in:
0.44880
outside
1 - 215
Population Genetic Test Statistics
Pi
243.964223
Theta
160.113104
Tajima's D
1.646816
CLR
0.000004
CSRT
0.814459277036148
Interpretation
Uncertain
