Pre Gene Modal
BGIBMGA007690
Annotation
serine/threonine_protein_phosphatase_6_[Bombyx_mori]
Full name
Serine/threonine-protein phosphatase
+ More
Serine/threonine-protein phosphatase 6 catalytic subunit
Serine/threonine-protein phosphatase 6 catalytic subunit
Alternative Name
Phosphatase V
Protein phosphatase V
Protein phosphatase V
Location in the cell
Cytoplasmic Reliability : 2.732
Sequence
CDS
ATGACGGATATAGATAAATGGATAGAAGTTGCAAAGAGATGCAAATACCTCCCGGAAGATGACCTACGGGAACTGTGCAATATAGTTTGTGATTTGTTACTTGAAGAACCTAATGTTCAGCCGGTACAGACACCTGTGACGGTTTGTGGTGATATTCATGGACAGTTCTATGATTTAGAGGAGCTTTTTCACATAGGTGGTCAAGTGCCATATACAAAATACATATTTATGGGAGATTATGTGGATCGAGGATATTATAGTTTAGAAACACTAACCTTGCTCATGGCCTTGAAAGCTAGATATCCAGATAGGATAATCTTATTAAGGGGTAATCATGAAACATGTCAAATAACTAAGGTTTATGGATTTTATGATGAATGTCTCAATAAATATGGAAATGCAAATGCATGGAAAGACTGTTGCAGAGTTTTTGATTTGCTCACTGTTGCTGCATTAATTGATGAAACCGTCCTATGTGTGCACGGTGGTCTATCACCAGAAATATCTATGATTGATCAAATAAGATGCATTGATAGAAATCAACAAATTCCACACAAAGGTGCATTCTGTGATTTGTTGTGGTCAGATCCTGCTGATGTGAAAATGTGGTCAGTAAGTCCTCGTGGAGCTGGATGGTTGTTTGGCTGTCAGGTTACAGAACTCTTCATGAATTACAATGATTTAACTCTTATATGCAGGGCACATCAATTAGTGAATGATGGTTATAAATACATGTTTGACAAAAGATTAGTAACAGTCTGGTCGGCGCCTAACTACTGTTACCGATGTGGTAATGTTGCCTCAATCTTGGAATTCAACACTGTTCATGATAGAACAGCAAGGTTATTCCAAGCAGTGCCTGACACCGAACGAGAAGTTCCTCCACAGCATACTACTCCATATTTCCTTTGA
Protein
MTDIDKWIEVAKRCKYLPEDDLRELCNIVCDLLLEEPNVQPVQTPVTVCGDIHGQFYDLEELFHIGGQVPYTKYIFMGDYVDRGYYSLETLTLLMALKARYPDRIILLRGNHETCQITKVYGFYDECLNKYGNANAWKDCCRVFDLLTVAALIDETVLCVHGGLSPEISMIDQIRCIDRNQQIPHKGAFCDLLWSDPADVKMWSVSPRGAGWLFGCQVTELFMNYNDLTLICRAHQLVNDGYKYMFDKRLVTVWSAPNYCYRCGNVASILEFNTVHDRTARLFQAVPDTEREVPPQHTTPYFL
Summary
Description
May be involved in controlling cellularization or in regulating transcription of the genes involved in this process.
Catalytic subunit of protein phosphatase 6 (PP6). PP6 is a component of a signaling pathway regulating cell cycle progression in response to IL2 receptor stimulation. N-terminal domain restricts G1 to S phase progression in cancer cells, in part through control of cyclin D1. During mitosis, regulates spindle positioning. Downregulates MAP3K7 kinase activation of the IL1 signaling pathway by dephosphorylation of MAP3K7. Participates also in the innate immune defense against viruses by desphosphorylating RIG-I/DDX58, an essential step that triggers RIG-I/DDX58-mediated signaling activation.
Catalytic subunit of protein phosphatase 6 (PP6). PP6 is a component of a signaling pathway regulating cell cycle progression in response to IL2 receptor stimulation. N-terminal domain restricts G1 to S phase progression in cancer cells, in part through control of cyclin D1. During mitosis, regulates spindle positioning. Downregulates MAP3K7 kinase activation of the IL1 signaling pathway by dephosphorylation of MAP3K7. Participates also in the innate immune defense against viruses by desphosphorylating RIG-I/DDX58, an essential step that triggers RIG-I/DDX58-mediated signaling activation.
Catalytic Activity
H2O + O-phospho-L-threonyl-[protein] = L-threonyl-[protein] + phosphate
H2O + O-phospho-L-seryl-[protein] = L-seryl-[protein] + phosphate
H2O + O-phospho-L-seryl-[protein] = L-seryl-[protein] + phosphate
Cofactor
Mn(2+)
Subunit
Protein phosphatase 6 (PP6) holoenzyme is proposed to be a heterotrimeric complex formed by the catalytic subunit, a SAPS domain-containing subunit (PP6R) and an ankyrin repeat-domain containing regulatory subunit (ARS). Interacts with subunits PPP6R1, PPP6R2 and PPP6R3. Interacts with subunit ANKRD28. Interacts with IGBP1. Interacts with MAP3K7. Interacts with NFKBIE. Interacts with TRIM14 and WRNIP1; these interactions positively regulate the RIG-I/DDX58 signaling pathway.
Similarity
Belongs to the PPP phosphatase family.
Belongs to the PPP phosphatase family. PP-6 (PP-V) subfamily.
Belongs to the PPP phosphatase family. PP-6 (PP-V) subfamily.
Keywords
Complete proteome
Cytoplasm
Hydrolase
Manganese
Metal-binding
Protein phosphatase
Reference proteome
Acetylation
Cell cycle
Immunity
Innate immunity
Mitochondrion
Feature
chain Serine/threonine-protein phosphatase 6 catalytic subunit
Uniprot
Q1HPV3
I6LWZ6
A0A2H1X0P5
A0A3S2TDY9
S4PIT6
A0A194PWI0
+ More
W0FXW4 A0A1L8DM96 E2B693 A0A023GL49 G3MNN8 A0A0C9RUU7 A0A131XL30 A0A1E1X4M3 A0A131YIP6 L7M6Y3 A0A151IL65 A0A067RDI9 A0A151IYY2 F4W3W8 A0A195FY99 A0A158NUW3 A0A151XFZ4 A0A0A9Y4D3 A0A293LI51 A0A224YZ55 A0A023FJX2 A0A0J7L2N5 E2AA60 A0A2R5LIQ6 A0A1B6DZA0 A0A3L8DHW2 A0A023FV98 A0A154PL02 A0A194QMW1 A0A2S2NMM7 A0A0L7QPJ6 A0A2H8TCL1 C4WVH6 A0A0B6ZVP9 V5HV40 T1I943 A0A1L8DKD4 A0A0N0BCL0 A0A310SEM2 A0A151I2I4 A0A0V0GBS1 A0A069DR82 A0A2A3EAW9 A0A088A6S1 A0A023F5P2 A0A076FFM7 A0A2S2R1N2 B4N2L1 V9KQQ6 M9PGI3 Q27884 A0A401PA56 A0A401SBA3 B4L2G5 A0A3B4DJV5 H3BFY5 A0A3B4H9U1 A0A3Q3CKX9 T1JA19 B3MXY0 Q52L13 B4M7X9 A0A1W4W2P9 A0A3P8PKI0 A0A1S3M4T1 A0A060XDV4 A0A3B1IPY2 B3NXS4 A0A1A8GC15 A0A1A8AT17 A0A1S3KYE4 Q7ZUS7 Q28GT8 A0A3L7HC66 A0A3Q0DA58 Q64620 Q9CQR6 A0A232EWF6 K7IZQ1 A0A1A8KM57 A0A1A8UAN6 A0A1A8RBS7 A0A1A8M844 A0A1A8CY77 A0A1A8FTU2 A0A1A8SEX8 A0A0P7WS88 A0A3B3I5J2 A0A3S2PFM8 A0A3P9JSI5 A0A3P8YZ84 A0A3Q3NCS7 A0A3Q4GW39 I3JZ68
W0FXW4 A0A1L8DM96 E2B693 A0A023GL49 G3MNN8 A0A0C9RUU7 A0A131XL30 A0A1E1X4M3 A0A131YIP6 L7M6Y3 A0A151IL65 A0A067RDI9 A0A151IYY2 F4W3W8 A0A195FY99 A0A158NUW3 A0A151XFZ4 A0A0A9Y4D3 A0A293LI51 A0A224YZ55 A0A023FJX2 A0A0J7L2N5 E2AA60 A0A2R5LIQ6 A0A1B6DZA0 A0A3L8DHW2 A0A023FV98 A0A154PL02 A0A194QMW1 A0A2S2NMM7 A0A0L7QPJ6 A0A2H8TCL1 C4WVH6 A0A0B6ZVP9 V5HV40 T1I943 A0A1L8DKD4 A0A0N0BCL0 A0A310SEM2 A0A151I2I4 A0A0V0GBS1 A0A069DR82 A0A2A3EAW9 A0A088A6S1 A0A023F5P2 A0A076FFM7 A0A2S2R1N2 B4N2L1 V9KQQ6 M9PGI3 Q27884 A0A401PA56 A0A401SBA3 B4L2G5 A0A3B4DJV5 H3BFY5 A0A3B4H9U1 A0A3Q3CKX9 T1JA19 B3MXY0 Q52L13 B4M7X9 A0A1W4W2P9 A0A3P8PKI0 A0A1S3M4T1 A0A060XDV4 A0A3B1IPY2 B3NXS4 A0A1A8GC15 A0A1A8AT17 A0A1S3KYE4 Q7ZUS7 Q28GT8 A0A3L7HC66 A0A3Q0DA58 Q64620 Q9CQR6 A0A232EWF6 K7IZQ1 A0A1A8KM57 A0A1A8UAN6 A0A1A8RBS7 A0A1A8M844 A0A1A8CY77 A0A1A8FTU2 A0A1A8SEX8 A0A0P7WS88 A0A3B3I5J2 A0A3S2PFM8 A0A3P9JSI5 A0A3P8YZ84 A0A3Q3NCS7 A0A3Q4GW39 I3JZ68
EC Number
3.1.3.16
Pubmed
19121390
23622113
26354079
20798317
22216098
28049606
+ More
28503490 26830274 25576852 24845553 21719571 21347285 25401762 26823975 28797301 30249741 25765539 26334808 25474469 17994087 24402279 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 8223492 12537569 30297745 9215903 27762356 24755649 25329095 23594743 20431018 29704459 26319212 8077208 15489334 16141072 19468303 16769727 21183079 28648823 20075255 17554307 25069045 25186727
28503490 26830274 25576852 24845553 21719571 21347285 25401762 26823975 28797301 30249741 25765539 26334808 25474469 17994087 24402279 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 8223492 12537569 30297745 9215903 27762356 24755649 25329095 23594743 20431018 29704459 26319212 8077208 15489334 16141072 19468303 16769727 21183079 28648823 20075255 17554307 25069045 25186727
EMBL
BABH01002941
DQ443299
ABF51388.1
JF704119
AEG64801.1
ODYU01012545
+ More
SOQ58930.1 RSAL01000268 RVE43220.1 GAIX01000074 JAA92486.1 KQ459596 KPI95490.1 KF255602 AHF45929.1 GFDF01006589 JAV07495.1 GL445930 EFN88786.1 GBBM01001878 JAC33540.1 JO843489 AEO35106.1 GBYB01011425 GBYB01011426 JAG81192.1 JAG81193.1 GEFH01002195 JAP66386.1 GFAC01005212 JAT93976.1 GEDV01009384 JAP79173.1 GACK01005432 JAA59602.1 KQ977133 KYN05492.1 KK852705 KDR18025.1 KQ980744 KYN13708.1 GL887490 EGI71103.1 KQ981161 KYN45413.1 ADTU01026743 KQ982174 KYQ59309.1 GBHO01017631 GBRD01006034 GDHC01015246 JAG25973.1 JAG59787.1 JAQ03383.1 GFWV01002799 MAA27529.1 GFPF01011721 MAA22867.1 GBBK01003097 JAC21385.1 LBMM01001030 KMQ97047.1 GL438029 EFN69698.1 GGLE01005286 MBY09412.1 GEDC01011771 GEDC01008432 GEDC01006290 JAS25527.1 JAS28866.1 JAS31008.1 QOIP01000008 RLU19509.1 GBBL01001534 JAC25786.1 KQ434936 KZC11968.1 KQ461195 KPJ06679.1 GGMR01005821 MBY18440.1 KQ414819 KOC60479.1 GFXV01000024 MBW11829.1 ABLF02030852 AK341520 BAH71896.1 HACG01024985 CEK71850.1 GANP01002079 JAB82389.1 ACPB03003492 GFDF01007165 JAV06919.1 KQ435899 KOX69134.1 KQ760631 OAD59710.1 KQ976534 KYM81474.1 GECL01000784 JAP05340.1 GBGD01002306 JAC86583.1 KZ288312 PBC28442.1 GBBI01002225 JAC16487.1 KF516596 AII16500.1 GGMS01014477 MBY83680.1 CH963925 EDW78600.1 JW868154 AFP00672.1 AE014298 AGB95118.1 X75980 BT021230 AY119179 BFAA01006078 GCB70031.1 BEZZ01000173 GCC27666.1 CH933810 EDW06841.1 AFYH01007584 AFYH01007585 JH431980 CH902630 EDV38595.1 BC094109 CM004481 AAH94109.1 OCT65562.1 OCT65563.1 CH940653 EDW62896.1 FR904844 FR905261 CDQ72595.1 CDQ77783.1 CH954180 EDV47375.1 HAEC01001203 SBQ69280.1 HADY01019131 SBP57616.1 CR925807 CU468955 BC047847 BC075751 AAH47847.1 AAH75751.1 AAMC01117200 BC123920 CR761231 AAI23921.1 CAJ81892.1 RAZU01000237 RLQ63534.1 X77236 BC078747 AK002764 AK009104 AL928639 BC002223 NNAY01001870 OXU22679.1 HAEE01013308 SBR33358.1 HAEJ01003935 SBS44392.1 HAEH01015428 SBS02724.1 HAEF01011824 HAEG01011067 SBR52983.1 HADZ01020821 HAEA01001556 SBP84762.1 HAEB01015751 SBQ62278.1 HAEI01013646 SBS16115.1 JARO02006823 KPP64726.1 CM012448 RVE65573.1 AERX01003745
SOQ58930.1 RSAL01000268 RVE43220.1 GAIX01000074 JAA92486.1 KQ459596 KPI95490.1 KF255602 AHF45929.1 GFDF01006589 JAV07495.1 GL445930 EFN88786.1 GBBM01001878 JAC33540.1 JO843489 AEO35106.1 GBYB01011425 GBYB01011426 JAG81192.1 JAG81193.1 GEFH01002195 JAP66386.1 GFAC01005212 JAT93976.1 GEDV01009384 JAP79173.1 GACK01005432 JAA59602.1 KQ977133 KYN05492.1 KK852705 KDR18025.1 KQ980744 KYN13708.1 GL887490 EGI71103.1 KQ981161 KYN45413.1 ADTU01026743 KQ982174 KYQ59309.1 GBHO01017631 GBRD01006034 GDHC01015246 JAG25973.1 JAG59787.1 JAQ03383.1 GFWV01002799 MAA27529.1 GFPF01011721 MAA22867.1 GBBK01003097 JAC21385.1 LBMM01001030 KMQ97047.1 GL438029 EFN69698.1 GGLE01005286 MBY09412.1 GEDC01011771 GEDC01008432 GEDC01006290 JAS25527.1 JAS28866.1 JAS31008.1 QOIP01000008 RLU19509.1 GBBL01001534 JAC25786.1 KQ434936 KZC11968.1 KQ461195 KPJ06679.1 GGMR01005821 MBY18440.1 KQ414819 KOC60479.1 GFXV01000024 MBW11829.1 ABLF02030852 AK341520 BAH71896.1 HACG01024985 CEK71850.1 GANP01002079 JAB82389.1 ACPB03003492 GFDF01007165 JAV06919.1 KQ435899 KOX69134.1 KQ760631 OAD59710.1 KQ976534 KYM81474.1 GECL01000784 JAP05340.1 GBGD01002306 JAC86583.1 KZ288312 PBC28442.1 GBBI01002225 JAC16487.1 KF516596 AII16500.1 GGMS01014477 MBY83680.1 CH963925 EDW78600.1 JW868154 AFP00672.1 AE014298 AGB95118.1 X75980 BT021230 AY119179 BFAA01006078 GCB70031.1 BEZZ01000173 GCC27666.1 CH933810 EDW06841.1 AFYH01007584 AFYH01007585 JH431980 CH902630 EDV38595.1 BC094109 CM004481 AAH94109.1 OCT65562.1 OCT65563.1 CH940653 EDW62896.1 FR904844 FR905261 CDQ72595.1 CDQ77783.1 CH954180 EDV47375.1 HAEC01001203 SBQ69280.1 HADY01019131 SBP57616.1 CR925807 CU468955 BC047847 BC075751 AAH47847.1 AAH75751.1 AAMC01117200 BC123920 CR761231 AAI23921.1 CAJ81892.1 RAZU01000237 RLQ63534.1 X77236 BC078747 AK002764 AK009104 AL928639 BC002223 NNAY01001870 OXU22679.1 HAEE01013308 SBR33358.1 HAEJ01003935 SBS44392.1 HAEH01015428 SBS02724.1 HAEF01011824 HAEG01011067 SBR52983.1 HADZ01020821 HAEA01001556 SBP84762.1 HAEB01015751 SBQ62278.1 HAEI01013646 SBS16115.1 JARO02006823 KPP64726.1 CM012448 RVE65573.1 AERX01003745
Proteomes
UP000005204
UP000283053
UP000053268
UP000008237
UP000078542
UP000027135
+ More
UP000078492 UP000007755 UP000078541 UP000005205 UP000075809 UP000036403 UP000000311 UP000279307 UP000076502 UP000053240 UP000053825 UP000007819 UP000015103 UP000053105 UP000078540 UP000242457 UP000005203 UP000007798 UP000000803 UP000288216 UP000287033 UP000009192 UP000261440 UP000008672 UP000261460 UP000264840 UP000007801 UP000186698 UP000008792 UP000192223 UP000265100 UP000087266 UP000193380 UP000018467 UP000008711 UP000000437 UP000008143 UP000273346 UP000189706 UP000002494 UP000000589 UP000215335 UP000002358 UP000034805 UP000192224 UP000001038 UP000265180 UP000265200 UP000265140 UP000261640 UP000261580 UP000005207
UP000078492 UP000007755 UP000078541 UP000005205 UP000075809 UP000036403 UP000000311 UP000279307 UP000076502 UP000053240 UP000053825 UP000007819 UP000015103 UP000053105 UP000078540 UP000242457 UP000005203 UP000007798 UP000000803 UP000288216 UP000287033 UP000009192 UP000261440 UP000008672 UP000261460 UP000264840 UP000007801 UP000186698 UP000008792 UP000192223 UP000265100 UP000087266 UP000193380 UP000018467 UP000008711 UP000000437 UP000008143 UP000273346 UP000189706 UP000002494 UP000000589 UP000215335 UP000002358 UP000034805 UP000192224 UP000001038 UP000265180 UP000265200 UP000265140 UP000261640 UP000261580 UP000005207
Interpro
Gene 3D
ProteinModelPortal
Q1HPV3
I6LWZ6
A0A2H1X0P5
A0A3S2TDY9
S4PIT6
A0A194PWI0
+ More
W0FXW4 A0A1L8DM96 E2B693 A0A023GL49 G3MNN8 A0A0C9RUU7 A0A131XL30 A0A1E1X4M3 A0A131YIP6 L7M6Y3 A0A151IL65 A0A067RDI9 A0A151IYY2 F4W3W8 A0A195FY99 A0A158NUW3 A0A151XFZ4 A0A0A9Y4D3 A0A293LI51 A0A224YZ55 A0A023FJX2 A0A0J7L2N5 E2AA60 A0A2R5LIQ6 A0A1B6DZA0 A0A3L8DHW2 A0A023FV98 A0A154PL02 A0A194QMW1 A0A2S2NMM7 A0A0L7QPJ6 A0A2H8TCL1 C4WVH6 A0A0B6ZVP9 V5HV40 T1I943 A0A1L8DKD4 A0A0N0BCL0 A0A310SEM2 A0A151I2I4 A0A0V0GBS1 A0A069DR82 A0A2A3EAW9 A0A088A6S1 A0A023F5P2 A0A076FFM7 A0A2S2R1N2 B4N2L1 V9KQQ6 M9PGI3 Q27884 A0A401PA56 A0A401SBA3 B4L2G5 A0A3B4DJV5 H3BFY5 A0A3B4H9U1 A0A3Q3CKX9 T1JA19 B3MXY0 Q52L13 B4M7X9 A0A1W4W2P9 A0A3P8PKI0 A0A1S3M4T1 A0A060XDV4 A0A3B1IPY2 B3NXS4 A0A1A8GC15 A0A1A8AT17 A0A1S3KYE4 Q7ZUS7 Q28GT8 A0A3L7HC66 A0A3Q0DA58 Q64620 Q9CQR6 A0A232EWF6 K7IZQ1 A0A1A8KM57 A0A1A8UAN6 A0A1A8RBS7 A0A1A8M844 A0A1A8CY77 A0A1A8FTU2 A0A1A8SEX8 A0A0P7WS88 A0A3B3I5J2 A0A3S2PFM8 A0A3P9JSI5 A0A3P8YZ84 A0A3Q3NCS7 A0A3Q4GW39 I3JZ68
W0FXW4 A0A1L8DM96 E2B693 A0A023GL49 G3MNN8 A0A0C9RUU7 A0A131XL30 A0A1E1X4M3 A0A131YIP6 L7M6Y3 A0A151IL65 A0A067RDI9 A0A151IYY2 F4W3W8 A0A195FY99 A0A158NUW3 A0A151XFZ4 A0A0A9Y4D3 A0A293LI51 A0A224YZ55 A0A023FJX2 A0A0J7L2N5 E2AA60 A0A2R5LIQ6 A0A1B6DZA0 A0A3L8DHW2 A0A023FV98 A0A154PL02 A0A194QMW1 A0A2S2NMM7 A0A0L7QPJ6 A0A2H8TCL1 C4WVH6 A0A0B6ZVP9 V5HV40 T1I943 A0A1L8DKD4 A0A0N0BCL0 A0A310SEM2 A0A151I2I4 A0A0V0GBS1 A0A069DR82 A0A2A3EAW9 A0A088A6S1 A0A023F5P2 A0A076FFM7 A0A2S2R1N2 B4N2L1 V9KQQ6 M9PGI3 Q27884 A0A401PA56 A0A401SBA3 B4L2G5 A0A3B4DJV5 H3BFY5 A0A3B4H9U1 A0A3Q3CKX9 T1JA19 B3MXY0 Q52L13 B4M7X9 A0A1W4W2P9 A0A3P8PKI0 A0A1S3M4T1 A0A060XDV4 A0A3B1IPY2 B3NXS4 A0A1A8GC15 A0A1A8AT17 A0A1S3KYE4 Q7ZUS7 Q28GT8 A0A3L7HC66 A0A3Q0DA58 Q64620 Q9CQR6 A0A232EWF6 K7IZQ1 A0A1A8KM57 A0A1A8UAN6 A0A1A8RBS7 A0A1A8M844 A0A1A8CY77 A0A1A8FTU2 A0A1A8SEX8 A0A0P7WS88 A0A3B3I5J2 A0A3S2PFM8 A0A3P9JSI5 A0A3P8YZ84 A0A3Q3NCS7 A0A3Q4GW39 I3JZ68
PDB
3P71
E-value=1.01811e-94,
Score=883
Ontologies
GO
Topology
Subcellular location
Cytoplasm
Mitochondrion
Mitochondrion
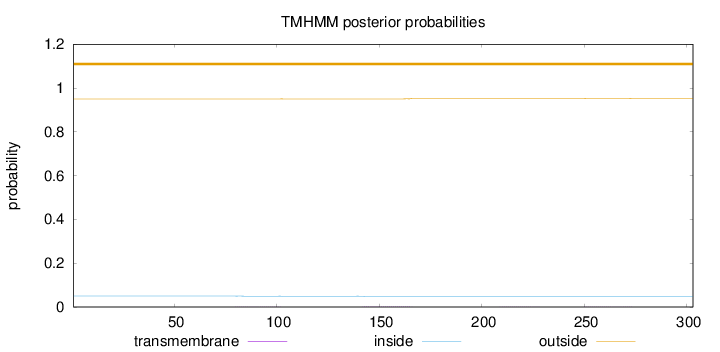
Length:
303
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0764
Exp number, first 60 AAs:
0
Total prob of N-in:
0.05008
outside
1 - 303
Population Genetic Test Statistics
Pi
121.894227
Theta
185.378363
Tajima's D
-1.101782
CLR
123.006367
CSRT
0.121893905304735
Interpretation
Uncertain
