Gene
KWMTBOMO08769
Pre Gene Modal
BGIBMGA007891
Annotation
PREDICTED:_GTPase-activating_Rap/Ran-GAP_domain-like_protein_3_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.32 Mitochondrial Reliability : 1.113 Nuclear Reliability : 1.102
Sequence
CDS
ATGCTGCCAACCTCCATTTCCGAATACGCCAACTCGTCGGCGAGTGAGCTGATAACAAGACGCGGCGTTTTTTCCAGACGACAATATGGTTCCGTTGAACAGTTATCAGGCACAAGCGACGGTGGTGGCGCTTTGGATGCAGTGGTGCGCCGTTACCGGCTCGAAAACGGAAACTCTCTCGGCGAAAAGGATGAATTGTTCGGACCGCCCAGCGCGCCCGTCCTTGAGAACCCTGAGTTCCAAACAAGATGGTACTTCAAATACTTTCTTGGTAAAATGCATCAGAATTACATCGGTGTCGATGGAGCGAGGGAGCCATATCTTCTCAGCGTTGTTCAGGAAGGGGGAGCAGGACTACTACGTGCCATACTTTTTAACAAAATGGGCGCACACAAAATATATTTGCCACCGAGTGCCTGGAGTGGAAGCGGTGGTCAAGCAACAAGGCCGACGGTAAAACAAATACTAGCCCAGTTTCCAGCTATGGAGAAAGTAGACAAAGTACCGAGAGAGATCACATGCGCCGAAATTCAGAAAGACGTTCTGCTGCTCGAAGAGCAAGAAGGCTCTGTGAACTTTAAAATAGGCGTGATGCTAATGAATCCAGGTCAGAAAACTGACGACGAGATGCTTTCGAACGAGAAGGGCGACGAGAAATGGGAACGATTTATTTCACTTTTGGGTGATAAAATTCGATTACGAGGATGGAACCGTTTCCGAGGCGGCCTCGACGTTAAAGGTGATATGACTGGCAGCCATTCCATTTACACGATGCATCAAGGACACGAAATTATGTTTCACGTCTCAACGATATTACCGTTTTCAAAGGATAACAAACAGCAGTTGGAAAGAAAACGGCATATTGGTAATGATATCGTGAATATAATTTTCTCGGATGACTCCGTCCACAATACCTTCAACCCGCAGTGCGTTAAAAGCCATTTCACGCATATTTTTGCCGTTGTATCTGAAGTGGAAGGAGCGGGATATAGGTTGAGTGTTTACAGCGATGACACGGTACCCCCATTTGGACCTTCGCTGCCTTGTCCCGCAATCTTCAGCGATCCCCAACTCTTCAGGGAATTTTTACTGGTGAAATTGATGAATGGAGAGAAAGCCGCGTTTCAAACTCCCACTTTTGCATTGAAACGACAAAGGACTCTTGATACATTGATAAGGGACATCTATGCAGAGCATTGTGCAGACCACAAAGTGCCCATGTTGTCACGACGCGCGCTCTCTGACGTCTGGTCCGGAGGTGCTGGGGCAGGAGCCGGGGCGGCGAGAACCGAAAGATTCTTACATGTTGGTCAAGCTCTTAAACTAGACGCGGTCTTAAGAGGCGACGCCCCAACCAGTCTTGTTTCTACAGGTTGCGGTGGCTTAAGACGTGCACCTTGGGAAGGTCGCGTGTGGAGAGCTGCTCCTCTTCCAGCATCTCCTGTTTGCGCTGAACAATTAGCAGAAGGACGACTACTCCTGTCTACTACCTCAAATACTTATATTTTAGAAGAAAGTGGAACTACACGCGTAGTCCTCAGATGGGAAGGTTGCGTGCGTGCGGCGCGTGTGAGCGAACGCGCTGGCTTGTTCCGCGTTGGAGCGCGTGTTGTGGCAGGCGGGGGCTCGACCACGGGAGGGGCCGGCGGTGCGGGATTGTACGCGCTACCACTGCCCGAACTCTGCGCCGGCGCCTGGGCAATGCGACCGCTGCAGGACTGTAAACACGCACGACTGCCGCGTACCAAGACCGCTCAATACTTCGATGTCCTGGAGTCAGTTAACGGAAATAAGACAACACTAGCAATAGCAGTCGGTAGAAGAGTGATGCTCATGATCGAAGAAGTTAAGGCTGACTATGAAATGGGATTCGAGTACGTACATATCAAGGACATTCAATTAAGCGAAACGCCGACCCTCATCAAGCTGATGGACGGTGAAGTTGAAACAATGGTGGTCGGCTACAAACACCATTGGGAGATTCTCGACACAAACAGTGGCCAAGGTGTGAAAAGAGCCGAAAATTCCGAAGACCCTGGAACCCGACGCGGGACTCTAGTCAATGCTCTGAGGATTGGCGACGAAAGAGACGGACAAATTGTATTATGTTACAATCATACATGTCACTTTGAGAGGCTGACAAGCAAAGGATTAGAGAGCGACGGAGAATATGACTTTCATTGGACCACAGTTCCTGCTGCTATCGTTTGTGCTTTTCCATACATAATGGCCTTCGCGGAAGATTTACTGGAGATACGATTAGTGGCAAACGGTTCTCTGGTGCACGCAGCCTGCATACCGGGCTTAAAGCTTCTGTGCGGACGAATGGATATATTCTACGTGGCCTGTGCTCCGGAATACGTACCATTGAGCCGTGAGATAGACCCGTGCTTGAGTCTGCACGACTACGAACTAGTCAAGCAGATGAACAAACAGAGCGGCCCGTATTCACTGGATGAAGATGAAAACCACGAAAATAAAGGCGACGCGGCTCATGCGAATCGAACATCCGAAGACAACTCAGCCTCGAGCAGAAGCAGCTCTATATCATCAGAACAGGAGAACATTAGGCCTCCCTTACAAAGGATGGCGCCTATATCACCGCCTACGTCACCCCAAGTAATGCCAGAGAATAGTCGTTGCGTGCGTATTTACAAAATACCACAGAGTAACCTCAGCGCGGGAACTTACCAAAGACAGTCCGTGTCAGCCGATCAAGTCGTCACCTCCTACTTTCCAGACAGACCGAACAGCATCAGACAGAGACTAGCGGAGTTGAGATTAGACAGCAAGTCTAGGGGAGGAGGCGATGACGAAAATTTGTGA
Protein
MLPTSISEYANSSASELITRRGVFSRRQYGSVEQLSGTSDGGGALDAVVRRYRLENGNSLGEKDELFGPPSAPVLENPEFQTRWYFKYFLGKMHQNYIGVDGAREPYLLSVVQEGGAGLLRAILFNKMGAHKIYLPPSAWSGSGGQATRPTVKQILAQFPAMEKVDKVPREITCAEIQKDVLLLEEQEGSVNFKIGVMLMNPGQKTDDEMLSNEKGDEKWERFISLLGDKIRLRGWNRFRGGLDVKGDMTGSHSIYTMHQGHEIMFHVSTILPFSKDNKQQLERKRHIGNDIVNIIFSDDSVHNTFNPQCVKSHFTHIFAVVSEVEGAGYRLSVYSDDTVPPFGPSLPCPAIFSDPQLFREFLLVKLMNGEKAAFQTPTFALKRQRTLDTLIRDIYAEHCADHKVPMLSRRALSDVWSGGAGAGAGAARTERFLHVGQALKLDAVLRGDAPTSLVSTGCGGLRRAPWEGRVWRAAPLPASPVCAEQLAEGRLLLSTTSNTYILEESGTTRVVLRWEGCVRAARVSERAGLFRVGARVVAGGGSTTGGAGGAGLYALPLPELCAGAWAMRPLQDCKHARLPRTKTAQYFDVLESVNGNKTTLAIAVGRRVMLMIEEVKADYEMGFEYVHIKDIQLSETPTLIKLMDGEVETMVVGYKHHWEILDTNSGQGVKRAENSEDPGTRRGTLVNALRIGDERDGQIVLCYNHTCHFERLTSKGLESDGEYDFHWTTVPAAIVCAFPYIMAFAEDLLEIRLVANGSLVHAACIPGLKLLCGRMDIFYVACAPEYVPLSREIDPCLSLHDYELVKQMNKQSGPYSLDEDENHENKGDAAHANRTSEDNSASSRSSSISSEQENIRPPLQRMAPISPPTSPQVMPENSRCVRIYKIPQSNLSAGTYQRQSVSADQVVTSYFPDRPNSIRQRLAELRLDSKSRGGGDDENL
Summary
Uniprot
A0A3S2LGF1
A0A2A4K826
A0A212F5N3
A0A194QNX4
A0A194PPW4
A0A2H1WSL7
+ More
A0A0L7LGC1 D6X4H2 A0A1Y1LP53 N6UBU0 A0A1S4GFY5 A0A1B6CQF3 A0A1A9YF06 A0A1A9W663 A0A0N8CXN9 A0A0P5U122 A0A0P5MR75 A0A0P5XI86 A0A0P5BT37 A0A0P5APE2 A0A0P5HCH8 A0A0P6G3Y6 A0A0N7ZMD5 A0A0P5BUP2 A0A182NRK1 A0A182K2M4 A0A182P074 A0A182Y6G1 A0A2R5L7B5 A0A0N8B8Y4 A0A0P5DG54 A0A0P5ZU72 A0A0P5MN85 A0A0N8CF21 A0A0P5JLY8 A0A0P5N1Z7 A0A0P5DDN9 A0A182FNV2 A0A0P5YBK6 A0A0P5WIK0 A0A0P5DNG2 A0A0P4ZZ10 A0A0P4YLY3 A0A0P5APE8 A0A0P4ZU07 A0A0P6C9K7 A0A0P4YQY5 A0A0P5E9M7 A0A0P5ZEZ8 A0A0N8AV79 A0A0P5UFD6 A0A0P5I1D6 A0A0P5AS00 A0A0P5EM46 A0A0P4Z8S0 A0A0P5GJ39 A0A0P6G2Y6 A0A0P5GQ29 A0A0P5MWL3 A0A0P5H3G5 A0A0P5N9C6 A0A0P5GU18 A0A0P5C3U6 A0A0P5CQJ2 A0A0P5GPY1 A0A0P5Y353 A0A0L7R3H3 W8AJE0 W8ARH1 A0A0N8D6A8 A0A0N8ENH6 A0A0N8AS84 A0A0P5E442 A0A131XWH6 A0A0P5KDG2 A0A0P5XCV1 A0A1E1X2G8 A0A147BU33 A0A195BSP9 A0A224YZV7 L7MIC4 A0A131YVM2 A0A224YYD8 L7M290 L7MIV6 L7LV71 A0A084WRI6 A0A131YCS5 A0A131XDQ9 A0A0C9R0T7 A0A310SPI2 A0A0P5IPC8 A0A067R3L1 A0A023F5M3 A0A293LI75 A0A0C9RSB3 A0A1B0A0Z0 A0A1E1XMI0 A0A151XAU7
A0A0L7LGC1 D6X4H2 A0A1Y1LP53 N6UBU0 A0A1S4GFY5 A0A1B6CQF3 A0A1A9YF06 A0A1A9W663 A0A0N8CXN9 A0A0P5U122 A0A0P5MR75 A0A0P5XI86 A0A0P5BT37 A0A0P5APE2 A0A0P5HCH8 A0A0P6G3Y6 A0A0N7ZMD5 A0A0P5BUP2 A0A182NRK1 A0A182K2M4 A0A182P074 A0A182Y6G1 A0A2R5L7B5 A0A0N8B8Y4 A0A0P5DG54 A0A0P5ZU72 A0A0P5MN85 A0A0N8CF21 A0A0P5JLY8 A0A0P5N1Z7 A0A0P5DDN9 A0A182FNV2 A0A0P5YBK6 A0A0P5WIK0 A0A0P5DNG2 A0A0P4ZZ10 A0A0P4YLY3 A0A0P5APE8 A0A0P4ZU07 A0A0P6C9K7 A0A0P4YQY5 A0A0P5E9M7 A0A0P5ZEZ8 A0A0N8AV79 A0A0P5UFD6 A0A0P5I1D6 A0A0P5AS00 A0A0P5EM46 A0A0P4Z8S0 A0A0P5GJ39 A0A0P6G2Y6 A0A0P5GQ29 A0A0P5MWL3 A0A0P5H3G5 A0A0P5N9C6 A0A0P5GU18 A0A0P5C3U6 A0A0P5CQJ2 A0A0P5GPY1 A0A0P5Y353 A0A0L7R3H3 W8AJE0 W8ARH1 A0A0N8D6A8 A0A0N8ENH6 A0A0N8AS84 A0A0P5E442 A0A131XWH6 A0A0P5KDG2 A0A0P5XCV1 A0A1E1X2G8 A0A147BU33 A0A195BSP9 A0A224YZV7 L7MIC4 A0A131YVM2 A0A224YYD8 L7M290 L7MIV6 L7LV71 A0A084WRI6 A0A131YCS5 A0A131XDQ9 A0A0C9R0T7 A0A310SPI2 A0A0P5IPC8 A0A067R3L1 A0A023F5M3 A0A293LI75 A0A0C9RSB3 A0A1B0A0Z0 A0A1E1XMI0 A0A151XAU7
Pubmed
EMBL
RSAL01000014
RVE53272.1
NWSH01000054
PCG80156.1
AGBW02010169
OWR49038.1
+ More
KQ461195 KPJ06675.1 KQ459596 KPI95486.1 ODYU01010694 SOQ55972.1 JTDY01001315 KOB74231.1 KQ971380 EEZ97268.2 GEZM01054824 JAV73376.1 APGK01041016 KB740987 ENN76122.1 AAAB01008859 GEDC01021539 JAS15759.1 GDIP01088735 JAM14980.1 GDIP01119272 JAL84442.1 GDIQ01152504 JAK99221.1 GDIP01071828 JAM31887.1 GDIP01195315 JAJ28087.1 GDIP01196367 JAJ27035.1 GDIQ01229780 JAK21945.1 GDIQ01057346 JAN37391.1 GDIP01231257 JAI92144.1 GDIP01180364 JAJ43038.1 GGLE01001252 MBY05378.1 GDIQ01201187 JAK50538.1 GDIP01162213 JAJ61189.1 GDIP01039438 GDIQ01030324 JAM64277.1 JAN64413.1 GDIQ01161054 GDIQ01136421 GDIP01089833 JAK90671.1 JAM13882.1 GDIP01138079 JAL65635.1 GDIQ01201188 JAK50537.1 GDIQ01152453 JAK99272.1 GDIP01157871 JAJ65531.1 GDIQ01152455 GDIP01062033 JAK99270.1 JAM41682.1 GDIP01085526 JAM18189.1 GDIP01157215 JAJ66187.1 GDIP01206216 JAJ17186.1 GDIP01225912 JAI97489.1 GDIP01196368 JAJ27034.1 GDIP01207916 JAJ15486.1 GDIP01003769 JAM99946.1 GDIP01225911 JAI97490.1 GDIP01163068 JAJ60334.1 GDIP01044585 GDIQ01039251 JAM59130.1 JAN55486.1 GDIQ01239627 JAK12098.1 GDIP01113788 JAL89926.1 GDIQ01242631 JAK09094.1 GDIP01195316 JAJ28086.1 GDIQ01267447 JAJ84277.1 GDIP01216410 JAJ06992.1 GDIQ01243872 JAK07853.1 GDIQ01039250 JAN55487.1 GDIQ01241230 JAK10495.1 GDIQ01152454 JAK99271.1 GDIQ01239628 JAK12097.1 GDIQ01152505 JAK99220.1 GDIQ01242630 JAK09095.1 GDIP01176368 JAJ47034.1 GDIP01167066 GDIQ01044356 JAJ56336.1 JAN50381.1 GDIQ01238038 JAK13687.1 GDIP01064582 JAM39133.1 KQ414663 KOC65369.1 GAMC01017920 JAB88635.1 GAMC01017918 JAB88637.1 GDIP01064583 JAM39132.1 GDIQ01009460 JAN85277.1 GDIQ01247987 JAK03738.1 GDIP01147327 JAJ76075.1 GEFM01004397 JAP71399.1 GDIQ01211536 JAK40189.1 GDIP01075576 JAM28139.1 GFAC01005741 JAT93447.1 GEGO01001085 JAR94319.1 KQ976419 KYM89307.1 GFPF01008196 MAA19342.1 GACK01001487 JAA63547.1 GEDV01005284 JAP83273.1 GFPF01008197 MAA19343.1 GACK01006884 JAA58150.1 GACK01001940 JAA63094.1 GACK01009532 JAA55502.1 ATLV01026094 ATLV01026095 ATLV01026096 KE525406 KFB52830.1 GEDV01012209 JAP76348.1 GEFH01002867 JAP65714.1 GBYB01004921 GBYB01006527 GBYB01010067 JAG74688.1 JAG76294.1 JAG79834.1 KQ761976 OAD56430.1 GDIQ01211537 JAK40188.1 KK852927 KDR13691.1 GBBI01002250 JAC16462.1 GFWV01002819 MAA27549.1 GBYB01010276 JAG80043.1 GFAA01003011 JAU00424.1 KQ982335 KYQ57511.1
KQ461195 KPJ06675.1 KQ459596 KPI95486.1 ODYU01010694 SOQ55972.1 JTDY01001315 KOB74231.1 KQ971380 EEZ97268.2 GEZM01054824 JAV73376.1 APGK01041016 KB740987 ENN76122.1 AAAB01008859 GEDC01021539 JAS15759.1 GDIP01088735 JAM14980.1 GDIP01119272 JAL84442.1 GDIQ01152504 JAK99221.1 GDIP01071828 JAM31887.1 GDIP01195315 JAJ28087.1 GDIP01196367 JAJ27035.1 GDIQ01229780 JAK21945.1 GDIQ01057346 JAN37391.1 GDIP01231257 JAI92144.1 GDIP01180364 JAJ43038.1 GGLE01001252 MBY05378.1 GDIQ01201187 JAK50538.1 GDIP01162213 JAJ61189.1 GDIP01039438 GDIQ01030324 JAM64277.1 JAN64413.1 GDIQ01161054 GDIQ01136421 GDIP01089833 JAK90671.1 JAM13882.1 GDIP01138079 JAL65635.1 GDIQ01201188 JAK50537.1 GDIQ01152453 JAK99272.1 GDIP01157871 JAJ65531.1 GDIQ01152455 GDIP01062033 JAK99270.1 JAM41682.1 GDIP01085526 JAM18189.1 GDIP01157215 JAJ66187.1 GDIP01206216 JAJ17186.1 GDIP01225912 JAI97489.1 GDIP01196368 JAJ27034.1 GDIP01207916 JAJ15486.1 GDIP01003769 JAM99946.1 GDIP01225911 JAI97490.1 GDIP01163068 JAJ60334.1 GDIP01044585 GDIQ01039251 JAM59130.1 JAN55486.1 GDIQ01239627 JAK12098.1 GDIP01113788 JAL89926.1 GDIQ01242631 JAK09094.1 GDIP01195316 JAJ28086.1 GDIQ01267447 JAJ84277.1 GDIP01216410 JAJ06992.1 GDIQ01243872 JAK07853.1 GDIQ01039250 JAN55487.1 GDIQ01241230 JAK10495.1 GDIQ01152454 JAK99271.1 GDIQ01239628 JAK12097.1 GDIQ01152505 JAK99220.1 GDIQ01242630 JAK09095.1 GDIP01176368 JAJ47034.1 GDIP01167066 GDIQ01044356 JAJ56336.1 JAN50381.1 GDIQ01238038 JAK13687.1 GDIP01064582 JAM39133.1 KQ414663 KOC65369.1 GAMC01017920 JAB88635.1 GAMC01017918 JAB88637.1 GDIP01064583 JAM39132.1 GDIQ01009460 JAN85277.1 GDIQ01247987 JAK03738.1 GDIP01147327 JAJ76075.1 GEFM01004397 JAP71399.1 GDIQ01211536 JAK40189.1 GDIP01075576 JAM28139.1 GFAC01005741 JAT93447.1 GEGO01001085 JAR94319.1 KQ976419 KYM89307.1 GFPF01008196 MAA19342.1 GACK01001487 JAA63547.1 GEDV01005284 JAP83273.1 GFPF01008197 MAA19343.1 GACK01006884 JAA58150.1 GACK01001940 JAA63094.1 GACK01009532 JAA55502.1 ATLV01026094 ATLV01026095 ATLV01026096 KE525406 KFB52830.1 GEDV01012209 JAP76348.1 GEFH01002867 JAP65714.1 GBYB01004921 GBYB01006527 GBYB01010067 JAG74688.1 JAG76294.1 JAG79834.1 KQ761976 OAD56430.1 GDIQ01211537 JAK40188.1 KK852927 KDR13691.1 GBBI01002250 JAC16462.1 GFWV01002819 MAA27549.1 GBYB01010276 JAG80043.1 GFAA01003011 JAU00424.1 KQ982335 KYQ57511.1
Proteomes
SUPFAM
SSF111347
SSF111347
Gene 3D
ProteinModelPortal
A0A3S2LGF1
A0A2A4K826
A0A212F5N3
A0A194QNX4
A0A194PPW4
A0A2H1WSL7
+ More
A0A0L7LGC1 D6X4H2 A0A1Y1LP53 N6UBU0 A0A1S4GFY5 A0A1B6CQF3 A0A1A9YF06 A0A1A9W663 A0A0N8CXN9 A0A0P5U122 A0A0P5MR75 A0A0P5XI86 A0A0P5BT37 A0A0P5APE2 A0A0P5HCH8 A0A0P6G3Y6 A0A0N7ZMD5 A0A0P5BUP2 A0A182NRK1 A0A182K2M4 A0A182P074 A0A182Y6G1 A0A2R5L7B5 A0A0N8B8Y4 A0A0P5DG54 A0A0P5ZU72 A0A0P5MN85 A0A0N8CF21 A0A0P5JLY8 A0A0P5N1Z7 A0A0P5DDN9 A0A182FNV2 A0A0P5YBK6 A0A0P5WIK0 A0A0P5DNG2 A0A0P4ZZ10 A0A0P4YLY3 A0A0P5APE8 A0A0P4ZU07 A0A0P6C9K7 A0A0P4YQY5 A0A0P5E9M7 A0A0P5ZEZ8 A0A0N8AV79 A0A0P5UFD6 A0A0P5I1D6 A0A0P5AS00 A0A0P5EM46 A0A0P4Z8S0 A0A0P5GJ39 A0A0P6G2Y6 A0A0P5GQ29 A0A0P5MWL3 A0A0P5H3G5 A0A0P5N9C6 A0A0P5GU18 A0A0P5C3U6 A0A0P5CQJ2 A0A0P5GPY1 A0A0P5Y353 A0A0L7R3H3 W8AJE0 W8ARH1 A0A0N8D6A8 A0A0N8ENH6 A0A0N8AS84 A0A0P5E442 A0A131XWH6 A0A0P5KDG2 A0A0P5XCV1 A0A1E1X2G8 A0A147BU33 A0A195BSP9 A0A224YZV7 L7MIC4 A0A131YVM2 A0A224YYD8 L7M290 L7MIV6 L7LV71 A0A084WRI6 A0A131YCS5 A0A131XDQ9 A0A0C9R0T7 A0A310SPI2 A0A0P5IPC8 A0A067R3L1 A0A023F5M3 A0A293LI75 A0A0C9RSB3 A0A1B0A0Z0 A0A1E1XMI0 A0A151XAU7
A0A0L7LGC1 D6X4H2 A0A1Y1LP53 N6UBU0 A0A1S4GFY5 A0A1B6CQF3 A0A1A9YF06 A0A1A9W663 A0A0N8CXN9 A0A0P5U122 A0A0P5MR75 A0A0P5XI86 A0A0P5BT37 A0A0P5APE2 A0A0P5HCH8 A0A0P6G3Y6 A0A0N7ZMD5 A0A0P5BUP2 A0A182NRK1 A0A182K2M4 A0A182P074 A0A182Y6G1 A0A2R5L7B5 A0A0N8B8Y4 A0A0P5DG54 A0A0P5ZU72 A0A0P5MN85 A0A0N8CF21 A0A0P5JLY8 A0A0P5N1Z7 A0A0P5DDN9 A0A182FNV2 A0A0P5YBK6 A0A0P5WIK0 A0A0P5DNG2 A0A0P4ZZ10 A0A0P4YLY3 A0A0P5APE8 A0A0P4ZU07 A0A0P6C9K7 A0A0P4YQY5 A0A0P5E9M7 A0A0P5ZEZ8 A0A0N8AV79 A0A0P5UFD6 A0A0P5I1D6 A0A0P5AS00 A0A0P5EM46 A0A0P4Z8S0 A0A0P5GJ39 A0A0P6G2Y6 A0A0P5GQ29 A0A0P5MWL3 A0A0P5H3G5 A0A0P5N9C6 A0A0P5GU18 A0A0P5C3U6 A0A0P5CQJ2 A0A0P5GPY1 A0A0P5Y353 A0A0L7R3H3 W8AJE0 W8ARH1 A0A0N8D6A8 A0A0N8ENH6 A0A0N8AS84 A0A0P5E442 A0A131XWH6 A0A0P5KDG2 A0A0P5XCV1 A0A1E1X2G8 A0A147BU33 A0A195BSP9 A0A224YZV7 L7MIC4 A0A131YVM2 A0A224YYD8 L7M290 L7MIV6 L7LV71 A0A084WRI6 A0A131YCS5 A0A131XDQ9 A0A0C9R0T7 A0A310SPI2 A0A0P5IPC8 A0A067R3L1 A0A023F5M3 A0A293LI75 A0A0C9RSB3 A0A1B0A0Z0 A0A1E1XMI0 A0A151XAU7
PDB
1SRQ
E-value=1.12066e-48,
Score=491
Ontologies
GO
Topology
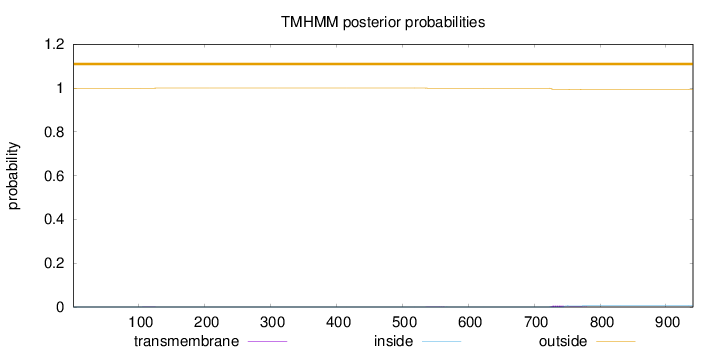
Length:
941
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.16286
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00043
outside
1 - 941
Population Genetic Test Statistics
Pi
288.831845
Theta
188.261446
Tajima's D
1.148521
CLR
0.508234
CSRT
0.697715114244288
Interpretation
Uncertain
