Pre Gene Modal
BGIBMGA007892
Annotation
Mitochondrial_ribosomal_protein_L30_[Operophtera_brumata]
Location in the cell
Cytoplasmic Reliability : 1.498 Mitochondrial Reliability : 1.579 Nuclear Reliability : 1.464
Sequence
CDS
ATGAACGGAAAATTACTACGAAATTTTAATCCACTGGTTTCTATCATTCGATCAAAAGGTTATAAACATCCAGGAGGTATTCGTTACCCTGGGGGCATAACATACTATCCCAGATTCCCAGATTACAAAGATCCTGAAATAACTCCATCAAAGTTGTTCAGAGTAGAGCGAATTAAATCAAGCAAACATTTTCCATACTGGCAAAAAAGGATATTGGATGAATTGAAAATTCATGAGGAGACCAGAGTCACTGTGGTTAAAAATATACCAGAAATCAATGCTAAACTATGGAAAATCAAGCATTTGATTAAAATTACACCAATTGAATTTCCTTATGGAGAACCAACTGCAGATGATATAAACTACACAATCCTTAAAGAAAATGGTCAATGCTTGGTTACAAAAAAATTGGAACCGGAATTCGGCCAGATCCAGGCCCTAGAAGAATTTGATAGTGATAGAAAAAAGATGAATTCTACAACCATAAAAAGAGATACTAGACATAGATGGAATACAGGATATGGAGGGGGATTTTAA
Protein
MNGKLLRNFNPLVSIIRSKGYKHPGGIRYPGGITYYPRFPDYKDPEITPSKLFRVERIKSSKHFPYWQKRILDELKIHEETRVTVVKNIPEINAKLWKIKHLIKITPIEFPYGEPTADDINYTILKENGQCLVTKKLEPEFGQIQALEEFDSDRKKMNSTTIKRDTRHRWNTGYGGGF
Summary
Similarity
Belongs to the TCP-1 chaperonin family.
Uniprot
H9JEE5
A0A0L7LFS8
A0A2A4K8Y5
A0A1E1WH02
A0A3S2NKI8
A0A2H1WXK3
+ More
A0A194QMB0 I4DL52 A0A212F5M7 A0A336M338 A0A3B0JY75 A0A1A9ZEK8 A0A1A9XBT0 A0A1I8NBC1 A0A1B0BDY2 A0A1L8DRH4 A0A1B0CCT7 W8C572 A0A1A9VCL6 B4R4J9 B4I0W9 B3MS73 A0A1B0FIV2 Q9W4I3 A0A034WHH0 A0A1W4U568 Q8SYI9 A0A1B0DRH4 A0A1I8PXK5 A0A0K8U7E3 B3NU50 A0A1A9WIM9 Q29G79 B4GW20 A0A1A9WZC4 B4JX57 B4Q037 A0A0L0CAI8 A0A1L8ED33 B4N291 B4MA81 A0A0K8TSD6 A0A0M3QZL7 A0A1L8ED32 B4L706 B4JX59 A0A1Y1KQ81 A0A1J1HZ82 U5EQI7 A0A0T6AWX3 A0A023EHE0 A0A182GMG7 D2A539 A0A1W4WXZ5 V5GLJ9 A0A1Q3FD20 Q17CV6 A0A182YA46 A0A1W4WN87 A0A2M3ZC77 B0WXT3 A0A2M3ZCT7 D6X4V9 A0A182WCV3 A0A182ITJ7 A0A2M4C0P3 A0A182RLN8 A0A2M4C0P2 A0A182Q2P9 A0A2M3ZCE9 A0A084WI48 T1DP19 W5J8U4 A0A182P8G0 A0A182MYE8 A0A182VN94 Q7Q8S3 A0A2M4AXB7 A0A2M4AWL7 J3JU30 A0A2M4AWQ0 A0A182FV10 A0A182K4Y6 A0A182X1T6 A0A182U1J6 A0A182HFQ1 A0A182LQ39 A0A2P8Z0C0 A0A1B6LYN7 A0A2S2QSE8 A0A0P4VW05 R4UM68 T1H9I4 A0A0V0G8Q3 E2AJD1 A0A069DPV9 A0A224XZP2 A0A023F1I1 A0A067QYE9 A0A2H8TZ04
A0A194QMB0 I4DL52 A0A212F5M7 A0A336M338 A0A3B0JY75 A0A1A9ZEK8 A0A1A9XBT0 A0A1I8NBC1 A0A1B0BDY2 A0A1L8DRH4 A0A1B0CCT7 W8C572 A0A1A9VCL6 B4R4J9 B4I0W9 B3MS73 A0A1B0FIV2 Q9W4I3 A0A034WHH0 A0A1W4U568 Q8SYI9 A0A1B0DRH4 A0A1I8PXK5 A0A0K8U7E3 B3NU50 A0A1A9WIM9 Q29G79 B4GW20 A0A1A9WZC4 B4JX57 B4Q037 A0A0L0CAI8 A0A1L8ED33 B4N291 B4MA81 A0A0K8TSD6 A0A0M3QZL7 A0A1L8ED32 B4L706 B4JX59 A0A1Y1KQ81 A0A1J1HZ82 U5EQI7 A0A0T6AWX3 A0A023EHE0 A0A182GMG7 D2A539 A0A1W4WXZ5 V5GLJ9 A0A1Q3FD20 Q17CV6 A0A182YA46 A0A1W4WN87 A0A2M3ZC77 B0WXT3 A0A2M3ZCT7 D6X4V9 A0A182WCV3 A0A182ITJ7 A0A2M4C0P3 A0A182RLN8 A0A2M4C0P2 A0A182Q2P9 A0A2M3ZCE9 A0A084WI48 T1DP19 W5J8U4 A0A182P8G0 A0A182MYE8 A0A182VN94 Q7Q8S3 A0A2M4AXB7 A0A2M4AWL7 J3JU30 A0A2M4AWQ0 A0A182FV10 A0A182K4Y6 A0A182X1T6 A0A182U1J6 A0A182HFQ1 A0A182LQ39 A0A2P8Z0C0 A0A1B6LYN7 A0A2S2QSE8 A0A0P4VW05 R4UM68 T1H9I4 A0A0V0G8Q3 E2AJD1 A0A069DPV9 A0A224XZP2 A0A023F1I1 A0A067QYE9 A0A2H8TZ04
Pubmed
19121390
26227816
26354079
22651552
22118469
25315136
+ More
24495485 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 15632085 17550304 26108605 26369729 28004739 24945155 26483478 18362917 19820115 17510324 25244985 24438588 20920257 23761445 12364791 14747013 17210077 22516182 23537049 20966253 29403074 27129103 20798317 26334808 25474469 24845553
24495485 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 15632085 17550304 26108605 26369729 28004739 24945155 26483478 18362917 19820115 17510324 25244985 24438588 20920257 23761445 12364791 14747013 17210077 22516182 23537049 20966253 29403074 27129103 20798317 26334808 25474469 24845553
EMBL
BABH01002965
JTDY01001315
KOB74229.1
NWSH01000054
PCG80158.1
PCG80160.1
+ More
GDQN01004771 JAT86283.1 RSAL01000014 RVE53268.1 ODYU01011831 SOQ57799.1 KQ461195 KPJ06673.1 AK402020 KQ459596 BAM18642.1 KPI95484.1 AGBW02010169 OWR49040.1 UFQT01000118 SSX20408.1 OUUW01000003 SPP77681.1 JXJN01012727 GFDF01005022 JAV09062.1 AJWK01007090 GAMC01001937 JAC04619.1 CM000366 EDX17089.1 CH480819 EDW53150.1 CH902622 EDV34628.1 CCAG010017454 AE014298 BT030799 AAF45968.1 ABV82181.1 GAKP01005352 JAC53600.1 AY071518 AAL49140.1 AJVK01020029 GDHF01029818 JAI22496.1 CH954180 EDV45826.1 CH379064 EAL32231.1 CH479193 EDW26865.1 CH916376 EDV95333.1 CM000162 EDX01189.1 JRES01000767 KNC28459.1 GFDG01002245 JAV16554.1 CH963925 EDW78480.1 CH940655 EDW66140.1 GDAI01000550 JAI17053.1 CP012528 ALC49599.1 GFDG01002243 JAV16556.1 CH933812 EDW06152.1 EDV95335.1 GEZM01081169 JAV61835.1 CVRI01000021 CRK91433.1 GANO01004263 JAB55608.1 LJIG01022640 KRT79492.1 GAPW01004800 JAC08798.1 JXUM01075592 KQ562895 KXJ74889.1 KQ971361 EFA05140.1 GALX01005994 JAB62472.1 GFDL01009657 JAV25388.1 CH477304 EAT44165.1 GGFM01005415 MBW26166.1 DS232173 EDS36743.1 GGFM01005537 MBW26288.1 KQ971381 EEZ97592.1 GGFJ01009732 MBW58873.1 GGFJ01009734 MBW58875.1 AXCN02001048 GGFM01005485 MBW26236.1 ATLV01023913 KE525347 KFB49892.1 GAMD01002345 JAA99245.1 ADMH02002040 ETN59803.1 AAAB01008933 EAA09955.3 GGFK01012090 MBW45411.1 GGFK01011667 MBW44988.1 APGK01043767 BT126742 KB741017 KB631644 AEE61704.1 ENN75254.1 ERL84939.1 GGFK01011811 MBW45132.1 APCN01005563 PYGN01000257 PSN49928.1 GEBQ01011184 JAT28793.1 GGMS01010849 MBY80052.1 GDKW01002776 JAI53819.1 KC632381 AGM32195.1 ACPB03006890 GECL01001718 JAP04406.1 GL439972 EFN66452.1 GBGD01003222 JAC85667.1 GFTR01002857 JAW13569.1 GBBI01003330 JAC15382.1 KK853197 KDR09964.1 GFXV01007691 MBW19496.1
GDQN01004771 JAT86283.1 RSAL01000014 RVE53268.1 ODYU01011831 SOQ57799.1 KQ461195 KPJ06673.1 AK402020 KQ459596 BAM18642.1 KPI95484.1 AGBW02010169 OWR49040.1 UFQT01000118 SSX20408.1 OUUW01000003 SPP77681.1 JXJN01012727 GFDF01005022 JAV09062.1 AJWK01007090 GAMC01001937 JAC04619.1 CM000366 EDX17089.1 CH480819 EDW53150.1 CH902622 EDV34628.1 CCAG010017454 AE014298 BT030799 AAF45968.1 ABV82181.1 GAKP01005352 JAC53600.1 AY071518 AAL49140.1 AJVK01020029 GDHF01029818 JAI22496.1 CH954180 EDV45826.1 CH379064 EAL32231.1 CH479193 EDW26865.1 CH916376 EDV95333.1 CM000162 EDX01189.1 JRES01000767 KNC28459.1 GFDG01002245 JAV16554.1 CH963925 EDW78480.1 CH940655 EDW66140.1 GDAI01000550 JAI17053.1 CP012528 ALC49599.1 GFDG01002243 JAV16556.1 CH933812 EDW06152.1 EDV95335.1 GEZM01081169 JAV61835.1 CVRI01000021 CRK91433.1 GANO01004263 JAB55608.1 LJIG01022640 KRT79492.1 GAPW01004800 JAC08798.1 JXUM01075592 KQ562895 KXJ74889.1 KQ971361 EFA05140.1 GALX01005994 JAB62472.1 GFDL01009657 JAV25388.1 CH477304 EAT44165.1 GGFM01005415 MBW26166.1 DS232173 EDS36743.1 GGFM01005537 MBW26288.1 KQ971381 EEZ97592.1 GGFJ01009732 MBW58873.1 GGFJ01009734 MBW58875.1 AXCN02001048 GGFM01005485 MBW26236.1 ATLV01023913 KE525347 KFB49892.1 GAMD01002345 JAA99245.1 ADMH02002040 ETN59803.1 AAAB01008933 EAA09955.3 GGFK01012090 MBW45411.1 GGFK01011667 MBW44988.1 APGK01043767 BT126742 KB741017 KB631644 AEE61704.1 ENN75254.1 ERL84939.1 GGFK01011811 MBW45132.1 APCN01005563 PYGN01000257 PSN49928.1 GEBQ01011184 JAT28793.1 GGMS01010849 MBY80052.1 GDKW01002776 JAI53819.1 KC632381 AGM32195.1 ACPB03006890 GECL01001718 JAP04406.1 GL439972 EFN66452.1 GBGD01003222 JAC85667.1 GFTR01002857 JAW13569.1 GBBI01003330 JAC15382.1 KK853197 KDR09964.1 GFXV01007691 MBW19496.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000283053
UP000053240
UP000053268
+ More
UP000007151 UP000268350 UP000092445 UP000092443 UP000095301 UP000092460 UP000092461 UP000078200 UP000000304 UP000001292 UP000007801 UP000092444 UP000000803 UP000192221 UP000092462 UP000095300 UP000008711 UP000091820 UP000001819 UP000008744 UP000001070 UP000002282 UP000037069 UP000007798 UP000008792 UP000092553 UP000009192 UP000183832 UP000069940 UP000249989 UP000007266 UP000192223 UP000008820 UP000076408 UP000002320 UP000075920 UP000075880 UP000075900 UP000075886 UP000030765 UP000000673 UP000075885 UP000075884 UP000075903 UP000007062 UP000019118 UP000030742 UP000069272 UP000075881 UP000076407 UP000075902 UP000075840 UP000075882 UP000245037 UP000015103 UP000000311 UP000027135
UP000007151 UP000268350 UP000092445 UP000092443 UP000095301 UP000092460 UP000092461 UP000078200 UP000000304 UP000001292 UP000007801 UP000092444 UP000000803 UP000192221 UP000092462 UP000095300 UP000008711 UP000091820 UP000001819 UP000008744 UP000001070 UP000002282 UP000037069 UP000007798 UP000008792 UP000092553 UP000009192 UP000183832 UP000069940 UP000249989 UP000007266 UP000192223 UP000008820 UP000076408 UP000002320 UP000075920 UP000075880 UP000075900 UP000075886 UP000030765 UP000000673 UP000075885 UP000075884 UP000075903 UP000007062 UP000019118 UP000030742 UP000069272 UP000075881 UP000076407 UP000075902 UP000075840 UP000075882 UP000245037 UP000015103 UP000000311 UP000027135
Interpro
IPR016082
Ribosomal_L30_ferredoxin-like
+ More
IPR005996 Ribosomal_L30_bac-type
IPR036919 L30_ferredoxin-like_sf
IPR002809 EMC3/TMCO1
IPR002423 Cpn60/TCP-1
IPR027410 TCP-1-like_intermed_sf
IPR017998 Chaperone_TCP-1
IPR002194 Chaperonin_TCP-1_CS
IPR012721 Chap_CCT_theta
IPR027413 GROEL-like_equatorial_sf
IPR027409 GroEL-like_apical_dom_sf
IPR005996 Ribosomal_L30_bac-type
IPR036919 L30_ferredoxin-like_sf
IPR002809 EMC3/TMCO1
IPR002423 Cpn60/TCP-1
IPR027410 TCP-1-like_intermed_sf
IPR017998 Chaperone_TCP-1
IPR002194 Chaperonin_TCP-1_CS
IPR012721 Chap_CCT_theta
IPR027413 GROEL-like_equatorial_sf
IPR027409 GroEL-like_apical_dom_sf
Gene 3D
ProteinModelPortal
H9JEE5
A0A0L7LFS8
A0A2A4K8Y5
A0A1E1WH02
A0A3S2NKI8
A0A2H1WXK3
+ More
A0A194QMB0 I4DL52 A0A212F5M7 A0A336M338 A0A3B0JY75 A0A1A9ZEK8 A0A1A9XBT0 A0A1I8NBC1 A0A1B0BDY2 A0A1L8DRH4 A0A1B0CCT7 W8C572 A0A1A9VCL6 B4R4J9 B4I0W9 B3MS73 A0A1B0FIV2 Q9W4I3 A0A034WHH0 A0A1W4U568 Q8SYI9 A0A1B0DRH4 A0A1I8PXK5 A0A0K8U7E3 B3NU50 A0A1A9WIM9 Q29G79 B4GW20 A0A1A9WZC4 B4JX57 B4Q037 A0A0L0CAI8 A0A1L8ED33 B4N291 B4MA81 A0A0K8TSD6 A0A0M3QZL7 A0A1L8ED32 B4L706 B4JX59 A0A1Y1KQ81 A0A1J1HZ82 U5EQI7 A0A0T6AWX3 A0A023EHE0 A0A182GMG7 D2A539 A0A1W4WXZ5 V5GLJ9 A0A1Q3FD20 Q17CV6 A0A182YA46 A0A1W4WN87 A0A2M3ZC77 B0WXT3 A0A2M3ZCT7 D6X4V9 A0A182WCV3 A0A182ITJ7 A0A2M4C0P3 A0A182RLN8 A0A2M4C0P2 A0A182Q2P9 A0A2M3ZCE9 A0A084WI48 T1DP19 W5J8U4 A0A182P8G0 A0A182MYE8 A0A182VN94 Q7Q8S3 A0A2M4AXB7 A0A2M4AWL7 J3JU30 A0A2M4AWQ0 A0A182FV10 A0A182K4Y6 A0A182X1T6 A0A182U1J6 A0A182HFQ1 A0A182LQ39 A0A2P8Z0C0 A0A1B6LYN7 A0A2S2QSE8 A0A0P4VW05 R4UM68 T1H9I4 A0A0V0G8Q3 E2AJD1 A0A069DPV9 A0A224XZP2 A0A023F1I1 A0A067QYE9 A0A2H8TZ04
A0A194QMB0 I4DL52 A0A212F5M7 A0A336M338 A0A3B0JY75 A0A1A9ZEK8 A0A1A9XBT0 A0A1I8NBC1 A0A1B0BDY2 A0A1L8DRH4 A0A1B0CCT7 W8C572 A0A1A9VCL6 B4R4J9 B4I0W9 B3MS73 A0A1B0FIV2 Q9W4I3 A0A034WHH0 A0A1W4U568 Q8SYI9 A0A1B0DRH4 A0A1I8PXK5 A0A0K8U7E3 B3NU50 A0A1A9WIM9 Q29G79 B4GW20 A0A1A9WZC4 B4JX57 B4Q037 A0A0L0CAI8 A0A1L8ED33 B4N291 B4MA81 A0A0K8TSD6 A0A0M3QZL7 A0A1L8ED32 B4L706 B4JX59 A0A1Y1KQ81 A0A1J1HZ82 U5EQI7 A0A0T6AWX3 A0A023EHE0 A0A182GMG7 D2A539 A0A1W4WXZ5 V5GLJ9 A0A1Q3FD20 Q17CV6 A0A182YA46 A0A1W4WN87 A0A2M3ZC77 B0WXT3 A0A2M3ZCT7 D6X4V9 A0A182WCV3 A0A182ITJ7 A0A2M4C0P3 A0A182RLN8 A0A2M4C0P2 A0A182Q2P9 A0A2M3ZCE9 A0A084WI48 T1DP19 W5J8U4 A0A182P8G0 A0A182MYE8 A0A182VN94 Q7Q8S3 A0A2M4AXB7 A0A2M4AWL7 J3JU30 A0A2M4AWQ0 A0A182FV10 A0A182K4Y6 A0A182X1T6 A0A182U1J6 A0A182HFQ1 A0A182LQ39 A0A2P8Z0C0 A0A1B6LYN7 A0A2S2QSE8 A0A0P4VW05 R4UM68 T1H9I4 A0A0V0G8Q3 E2AJD1 A0A069DPV9 A0A224XZP2 A0A023F1I1 A0A067QYE9 A0A2H8TZ04
PDB
6GB2
E-value=3.22272e-15,
Score=194
Ontologies
GO
PANTHER
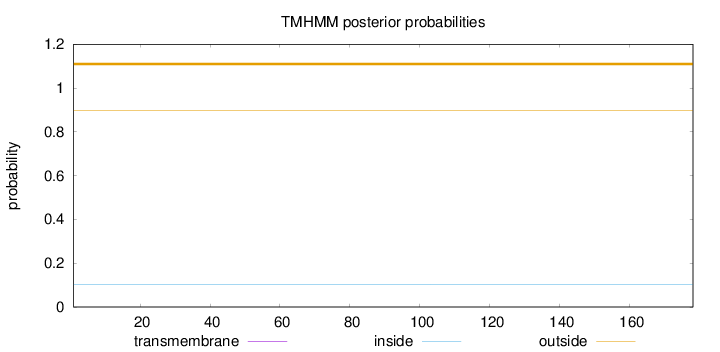
Topology
Subcellular location
Cytoplasm
Length:
178
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0002
Exp number, first 60 AAs:
0.0002
Total prob of N-in:
0.10243
outside
1 - 178
Population Genetic Test Statistics
Pi
38.48824
Theta
38.208816
Tajima's D
0.022645
CLR
0.637933
CSRT
0.380880955952202
Interpretation
Uncertain
