Gene
KWMTBOMO08765 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007893
Annotation
PREDICTED:_probable_E3_ubiquitin-protein_ligase_HERC2_[Amyelois_transitella]
Full name
E3 ubiquitin-protein ligase HERC2
Alternative Name
HECT domain and RCC1-like domain-containing protein 2
HECT-type E3 ubiquitin transferase HERC2
HECT-type E3 ubiquitin transferase HERC2
Location in the cell
Extracellular Reliability : 1.139 PlasmaMembrane Reliability : 1.439
Sequence
CDS
ATGTCAAGCCATGACGAGCACGAACAGTTCATTTTTCGTCCACAACAACATTTGGATGCTAAATGGCTCAATCCTGACTTACAGGAGGCCTTACACAATCCAGATTCCCTTTCTGCTTTATTTAATATTCTTTTACAAGACAGAGAAATATATTTCAATGCCGGAAGCGGCCTCGTAAACGGTGCTGGTATTACTGCTAAACTTGGGGATAGTGGAAAATACTACTGCGGCCTAAGGATATTAACCTGTACTTGTTGTGATGGTCTTTGTGGTCCCCACTCTGGATGTGCTTGTGTTCCATGCACTGCACTATCTGTTGAAGAAGAGCATAGAATATCTATCCTGACAAAATATGTGAATCCGCCAACATCACTTCAAATAATAGATGATCTCAAATGGAAACAAGATCCAGGACCAGAATGCCTTCTAAGTTTAATGGAGTCATTGATTTTTGAGCAAAGATTTAGAGCCATCAACACTGCTGTCAGCTGTCCATTAGTTTCAGAAATCCGTCATCTCATCGTCTTGTGCAACAGACATCTTGTGGCAGTTATAAGGGATCAAAGTCACAGAAAAGAAGTTAAAAAAGCGAATTTAGAGAAGAAACCTCCACATGGTGTAAGCAATAGAGCACTAAGCAATGTTGTAAATGCACTTAACAACCCCATAGATGCAAGCGCAGAAAGTCAAGAAGAAATTGAGGAGCCCACTGATGATTCAGAAAGTACTCTCGGCTTGGCACGGGTTGGTGCTCGTGCAGCGTTACGCTTAGCTTTGTCGCTGGTACGTCGTGCGTGGCGCTGTGGAGAAGATGCAGATGTTTGCTCAGCTTTGCTGAAAGATGCGTTGGATGCAGTGCGGGCTTTACCTGATGCTGCGCTATATGCCGGTACAGGGACCCCACAGCAAGCGCCAAGATCGCAGAAAATATGGGCGGAAGTTGTAGATAGCGCTGCTCAGTTCCTACATCAGGTTGTGACAGGTGAAGTGGGTTGTAATGTACCTGTTGGTGATTGGCGTATATCATTATGTGTGTGGGTTGAGTTGTGTGCGAGACGTGCCGAACTGCCAGCACTACTGAAAGCAGCTGATGTTCTAGTCACTTTACCTCCTAAGCAAAAACGACAGCCTGCCAATCACAGAATTACTCTAGAAGAATGTACAGCGCCCTTGGGGCCTTTTCTCAGACGTATGGCAAAAGTCGCAGCGCCTAACCCCATGGTCAACAATGAACCTTCAACAGAAACAAATCCCACTGAAGTATTTTTAAAAGAGCTGAATATTCCAAGCGGCGATGGTTTGATGAGTGTAAGAAAAGCGGGTATTGCGTTACTGTGTCATTTAGATAGACTCGGGGCACCCCTACTACCACCATTAAAAGGATTCGTAACGTGTACAGAATCCGCCCAAGAAGTGATCTCAGTAGGATTAGGCCCTATACAACTAGGTTCACTTAGAATACAGCAAATATCCTGTGCAGAAAAATTAGCATTACTTTTGACTCAAGACGGAGTTGTATACACACTGCCCTATGATACTATGACACCACAGTTGGTTCCAGGTTTGGAAAATAAGAGTATATGTCAAGTGGCATGTCACTCCGCCGGACGACACTTCTTGTGCTGCTCGTCGTGCGGCGGCGTGTGGTCCTGGGGTGTCGGCGAGGACGGCCGGCTCGGACACGGCGATACCGCTAATAGGGATGCGCCCTCTGCAATACATCACCTCGCACATCACGAAGTGACGCGCGTCACTGCCGGTGTGGCCTGCAGCGCTGTTATAACATCGCGTGGCGCTTTGTATACGTGGGGTCGTGGACCTCACGGCCGTCTCGGTCACGGCACTATTGAAAATTGTCTCACTCCACAACCCGTGTCCTTTAACGCGCATAACATTGAACGCATTATAGACGTGTCTTTAGGTTGGGGTGACAGCCTCACATTGTGTTGTACATGGGAGGGGGCAGTGCAAGGATTTGGCGACAATGAAGGCGGTGCTCCTAGAACCATAGCATCACTTTCGGTATTAAGGATCACAAGGGTATATACTGGAGAGCATTTCCACGCTGTTCTGACTGACAATGGTCAAGTTTATACATGGGGAAAAGGCGATAATTACCGTCTAGGACACGGCAACCTGGAAAACGTTAAGCTCCCAAAAGTAATGGAAGCTTTTCAAGGTATACGTATAATAGACGTGTCATTGGGATTATGTCACGGAGTAGCTCTGACGTCAGAGGGCGCCGTGTACGCTTGGGGCACGCACGAACGAGCGCAAATCGTGCGACCTGTACCGCAATTACTACAAGTACTCAGTCCTTCGTTTAAAGCGAATGGAATTAGCGGTGGTCCAAATCATATATTCGTATGGAGTGAAGAAAATCCCAGAGACTTACCATCGTCTATGCCTTTCGTGATAGATTTGTCAGATGGGACCTTTAAATTTTTAGAAAGACTTCTAAGATTAGTTGTGGAACAAAGTACATCAGGTGATCCAAAAGAGAATGAATGCTTAGCGGTAGCGTGTTTGAATCTACTGAGGTTGCAGGTTCATGCGTGGCAATACTCCCGAGGTATAGATTCGGTGGAGAGCGAGACAGAGAGTAGATCGTGGTGCGGCGGCGTGCACAGCGCACTGGTGTCGCTGGCGGAGGGGCGCGCGGGTCCTGTAGCCGCGGTGGCGGCGCGGCGCGCGCTGGCCGCGTCGTGGCCGCTGCTGCTGCCCACGCCTCAAGTTCGGGCACATCATCTCATTTCCTTATTGCCTCAAGGTTCAATGCCCGCTAGTGCAGGCGCACGTTTCATGACAGAGCTCCTGGTGTGGTCTCTGCTGGGGGGAGGAGGTCTACGTGACGCCGTACGGAATGCTTTGCAAATTCATTCAGCCGAGATGTATAACCATTTGCAGCTTGATAACGAATATGACCAGGTGGATATAATAACAGGACAACGTGGTCCTCCCGTATCGAAAACCGGCGACGATCCTGATCAGGTCACCGGGTTCTCTGAAGAATATCATCTGAGCAACACTGCAACATCGGACAAGTGCGCCTCCGTACCGCTCATGCACTTGGTGAAACATTTGATACGAAATACAAGCTGTCAGACACAAACGTCCATAAACGCAATAATTAATGGTGATCCCAGCAAGGTGGATGAGCCCAAGGGCCAAGACGCCGTCAGCCCCAGCTGTGATCTACTCATACGATTTCAAAGGTTATTGTTCTGTGAGGCGATGTGGTGTCGCGGCGGCCGCAGCTCGCGCGGCGGAGTCCGCTCGCTCCTGAACCAATACACGGTTCTACTGTCCGAGCACGTACAGTCGACGCTGAGCTCGTTCTCGCGAGCCGTGCACGGCGCCGACAATACCCAGCTCGTGGCTCTGTGTGATCTGCTAGCTTCTGACATTGTCGGTCGCGTTGTGACGGAGTTGAGCGCATGGATATGTGCTTGTTCGGCGCGGTCCGCACACGCGCTGTCCGACTGCGCGGGCTCGCTCGTGAGGATAGCGCGCGCGTGCGACGAGGCGGCCCGCGCGTTACCCTCGTCCGCGAAGCTAGACGCGGACCTGCTGGGCTGGCCCGGCCTCGTTTCGAAGAAACACCAGCTGAACAGTAGCGTTATACGCAAATGCGACTTACAGAATCATATAAAAGACGGCGGTTCGTGGATAATAGTGGCCGGTCACGTTTTCGACGTTGAGAATTTCGATTGCGAGAACGCGAGCACGGTGGAGTTGGTCCGCAAGTTGCGGGGCGGCGACGCGACCGCGGCCCTGAGCGTGGAGCCGCACGCCGCATTCCTAGCGCGGATCACTGAGAAGTGCGTCGGCCTCTATGCAGATCAGATTTATAGTCACAAATGTCATGAGAGTACGTCCAGTCTTCGTGCCCATCACATCCTGTCGTCTGTCGCGTTCCACGTCGCGGTGGGGCTGGCGCAGTGCGGCTCGAGGCTGGCGCGGTCGCTGCCCGTGCAGCAGGCGGAGCTCGACGCCGCGCCGTATGCAGCGGCTGTATTCCTGCGCGGGGGAATGCAGGCATATAGTGCGCGACGCGTCAATCCGAAACCGAGACCGTAA
Protein
MSSHDEHEQFIFRPQQHLDAKWLNPDLQEALHNPDSLSALFNILLQDREIYFNAGSGLVNGAGITAKLGDSGKYYCGLRILTCTCCDGLCGPHSGCACVPCTALSVEEEHRISILTKYVNPPTSLQIIDDLKWKQDPGPECLLSLMESLIFEQRFRAINTAVSCPLVSEIRHLIVLCNRHLVAVIRDQSHRKEVKKANLEKKPPHGVSNRALSNVVNALNNPIDASAESQEEIEEPTDDSESTLGLARVGARAALRLALSLVRRAWRCGEDADVCSALLKDALDAVRALPDAALYAGTGTPQQAPRSQKIWAEVVDSAAQFLHQVVTGEVGCNVPVGDWRISLCVWVELCARRAELPALLKAADVLVTLPPKQKRQPANHRITLEECTAPLGPFLRRMAKVAAPNPMVNNEPSTETNPTEVFLKELNIPSGDGLMSVRKAGIALLCHLDRLGAPLLPPLKGFVTCTESAQEVISVGLGPIQLGSLRIQQISCAEKLALLLTQDGVVYTLPYDTMTPQLVPGLENKSICQVACHSAGRHFLCCSSCGGVWSWGVGEDGRLGHGDTANRDAPSAIHHLAHHEVTRVTAGVACSAVITSRGALYTWGRGPHGRLGHGTIENCLTPQPVSFNAHNIERIIDVSLGWGDSLTLCCTWEGAVQGFGDNEGGAPRTIASLSVLRITRVYTGEHFHAVLTDNGQVYTWGKGDNYRLGHGNLENVKLPKVMEAFQGIRIIDVSLGLCHGVALTSEGAVYAWGTHERAQIVRPVPQLLQVLSPSFKANGISGGPNHIFVWSEENPRDLPSSMPFVIDLSDGTFKFLERLLRLVVEQSTSGDPKENECLAVACLNLLRLQVHAWQYSRGIDSVESETESRSWCGGVHSALVSLAEGRAGPVAAVAARRALAASWPLLLPTPQVRAHHLISLLPQGSMPASAGARFMTELLVWSLLGGGGLRDAVRNALQIHSAEMYNHLQLDNEYDQVDIITGQRGPPVSKTGDDPDQVTGFSEEYHLSNTATSDKCASVPLMHLVKHLIRNTSCQTQTSINAIINGDPSKVDEPKGQDAVSPSCDLLIRFQRLLFCEAMWCRGGRSSRGGVRSLLNQYTVLLSEHVQSTLSSFSRAVHGADNTQLVALCDLLASDIVGRVVTELSAWICACSARSAHALSDCAGSLVRIARACDEAARALPSSAKLDADLLGWPGLVSKKHQLNSSVIRKCDLQNHIKDGGSWIIVAGHVFDVENFDCENASTVELVRKLRGGDATAALSVEPHAAFLARITEKCVGLYADQIYSHKCHESTSSLRAHHILSSVAFHVAVGLAQCGSRLARSLPVQQAELDAAPYAAAVFLRGGMQAYSARRVNPKPRP
Summary
Description
E3 ubiquitin-protein ligase that regulates ubiquitin-dependent retention of repair proteins on damaged chromosomes. Recruited to sites of DNA damage in response to ionizing radiation (IR) and facilitates the assembly of UBE2N and RNF8 promoting DNA damage-induced formation of 'Lys-63'-linked ubiquitin chains. Acts as a mediator of binding specificity between UBE2N and RNF8. Involved in the maintenance of RNF168 levels. E3 ubiquitin-protein ligase that promotes the ubiquitination and proteasomal degradation of XPA which influences the circadian oscillation of DNA excision repair activity. By controlling the steady-state expression of the IGF1R receptor, indirectly regulates the insulin-like growth factor receptor signaling pathway (PubMed:26692333).
Catalytic Activity
S-ubiquitinyl-[E2 ubiquitin-conjugating enzyme]-L-cysteine + [acceptor protein]-L-lysine = [E2 ubiquitin-conjugating enzyme]-L-cysteine + N(6)-ubiquitinyl-[acceptor protein]-L-lysine.
Subunit
Interacts (when phosphorylated at Thr-4827 and sumoylated) with RNF8 (via FHA domain); this interaction increases after ionizing radiation (IR) treatment. Interacts with XPA. Interacts with NEURL4. Via its interaction with NEURL4, may indirectly interact with CCP110 and CEP97.
Miscellaneous
A regulatory element withinin an intron of the HERC2 gene inhibits OCA2 promoter. There are several single nucleotide polymorphisms within the OCA2 gene and within the HERC2 gene that have a statistical association with human eye color.
Keywords
3D-structure
Coiled coil
Complete proteome
Cytoplasm
Cytoskeleton
Disease mutation
DNA damage
DNA repair
Mental retardation
Metal-binding
Nucleus
Phosphoprotein
Reference proteome
Repeat
Transferase
Ubl conjugation
Ubl conjugation pathway
Zinc
Zinc-finger
Feature
chain E3 ubiquitin-protein ligase HERC2
sequence variant In MRT38; less stable than wild-type; diffuse cytosolic localization with the formation of abnormal aggregates; dbSNP:rs397518474.
sequence variant In MRT38; less stable than wild-type; diffuse cytosolic localization with the formation of abnormal aggregates; dbSNP:rs397518474.
Uniprot
H9JEE6
A0A2A4K8M6
A0A3S2PJI5
A0A2H1WXV6
A0A194PPV9
A0A026WJZ9
+ More
A0A3L8DD68 E2BM75 E2AK78 A0A0J7L4R0 A0A139WAN5 K7IQ42 A0A067R2N5 A0A158NCL1 F4WFF7 A0A151X600 A0A2A3E3G7 A0A195F9S1 A0A195ELX0 A0A195CR34 A0A088AL05 A0A139WAP8 A0A139WAR9 A0A154P7I9 A0A195BYI1 A0A146M433 V3ZE73 A0A1V4JBE1 H0ZJQ4 A0A2I0MLM1 E1BW48 G1KDV1 U3I746 U3JN73 A0A2T7PNV3 A0A091JXI0 A0A093HB52 A0A2I0MLL8 A0A091MCL6 A0A3M0K0M4 A0A3Q2UE20 R0L7U7 A0A091S499 A0A091JP83 A0A286XAB6 H0VHC2 A0A091EHF1 A0A091I9P4 A0A091LFD1 A0A093FPE5 I3M8T9 A0A287CWT3 A0A091VSZ3 A0A093CBX9 A0A0B8RTL9 F7A4S9 A0A384DC01 A0A091HM07 A0A061IGX1 A0A3Q0DBS9 A0A3Q7XI86 A0A094KRD9 A0A3Q2HE57 A0A3Q2IC42 F7AM44 A0A0Q3TAC1 A0A3Q2IA83 G3STX9 G5AZH7 A0A0P6J146 A0A2Y9QW94 E1B782 F7EP78 A0A0A0A8M2 F1RX57 A0A093BN38 A0A091TBI0 H9G1F0 A0A452GTE7 A0A452GSK3 A0A1U7UNG5 O95714 A0A250Y8J0 A0A151NXF6 A0A2K5K3Q5 A0A1U7RFM8 A0A287B1U2 A0A452GSM4 A0A3L8SV73 U3EA95 A0A2K5K3S5 A0A0G2JUC9 A0A452GSJ6 A0A2U4B2F2 A0A3Q7S9L4 A0A0D9R2F7 W5Q2G3
A0A3L8DD68 E2BM75 E2AK78 A0A0J7L4R0 A0A139WAN5 K7IQ42 A0A067R2N5 A0A158NCL1 F4WFF7 A0A151X600 A0A2A3E3G7 A0A195F9S1 A0A195ELX0 A0A195CR34 A0A088AL05 A0A139WAP8 A0A139WAR9 A0A154P7I9 A0A195BYI1 A0A146M433 V3ZE73 A0A1V4JBE1 H0ZJQ4 A0A2I0MLM1 E1BW48 G1KDV1 U3I746 U3JN73 A0A2T7PNV3 A0A091JXI0 A0A093HB52 A0A2I0MLL8 A0A091MCL6 A0A3M0K0M4 A0A3Q2UE20 R0L7U7 A0A091S499 A0A091JP83 A0A286XAB6 H0VHC2 A0A091EHF1 A0A091I9P4 A0A091LFD1 A0A093FPE5 I3M8T9 A0A287CWT3 A0A091VSZ3 A0A093CBX9 A0A0B8RTL9 F7A4S9 A0A384DC01 A0A091HM07 A0A061IGX1 A0A3Q0DBS9 A0A3Q7XI86 A0A094KRD9 A0A3Q2HE57 A0A3Q2IC42 F7AM44 A0A0Q3TAC1 A0A3Q2IA83 G3STX9 G5AZH7 A0A0P6J146 A0A2Y9QW94 E1B782 F7EP78 A0A0A0A8M2 F1RX57 A0A093BN38 A0A091TBI0 H9G1F0 A0A452GTE7 A0A452GSK3 A0A1U7UNG5 O95714 A0A250Y8J0 A0A151NXF6 A0A2K5K3Q5 A0A1U7RFM8 A0A287B1U2 A0A452GSM4 A0A3L8SV73 U3EA95 A0A2K5K3S5 A0A0G2JUC9 A0A452GSJ6 A0A2U4B2F2 A0A3Q7S9L4 A0A0D9R2F7 W5Q2G3
EC Number
2.3.2.26
Pubmed
19121390
26354079
24508170
30249741
20798317
18362917
+ More
19820115 20075255 24845553 21347285 21719571 26823975 23254933 20360741 23371554 15592404 23749191 21993624 20431018 23929341 26319212 19892987 21993625 19393038 17495919 30723633 25319552 28562605 9949213 16572171 10720573 17081983 18252221 18252222 18172690 18669648 19413330 19690332 20457063 20023648 20304803 20068231 21269460 21406692 22508508 22261722 23186163 26692333 23065719 28087693 22293439 30282656 25243066 15057822 22673903 20809919
19820115 20075255 24845553 21347285 21719571 26823975 23254933 20360741 23371554 15592404 23749191 21993624 20431018 23929341 26319212 19892987 21993625 19393038 17495919 30723633 25319552 28562605 9949213 16572171 10720573 17081983 18252221 18252222 18172690 18669648 19413330 19690332 20457063 20023648 20304803 20068231 21269460 21406692 22508508 22261722 23186163 26692333 23065719 28087693 22293439 30282656 25243066 15057822 22673903 20809919
EMBL
BABH01002966
BABH01002967
BABH01002968
BABH01002969
BABH01002970
BABH01002971
+ More
BABH01002972 NWSH01000054 PCG80163.1 RSAL01000014 RVE53266.1 ODYU01011831 SOQ57796.1 KQ459596 KPI95481.1 KK107167 EZA56377.1 QOIP01000010 RLU17868.1 GL449158 EFN83201.1 GL440194 EFN66182.1 LBMM01000739 KMQ97601.1 KQ971380 KYB24996.1 KK852747 KDR17278.1 ADTU01011956 ADTU01011957 GL888115 EGI67208.1 KQ982490 KYQ55797.1 KZ288445 PBC25641.1 KQ981727 KYN36962.1 KQ978691 KYN29213.1 KQ977408 KYN02952.1 KYB24998.1 KYB24997.1 KQ434831 KZC07832.1 KQ976396 KYM92996.1 GDHC01005102 JAQ13527.1 KB202591 ESO89418.1 LSYS01008075 OPJ69496.1 ABQF01013058 AKCR02000007 PKK30566.1 AADN05000806 ADON01107485 AGTO01010249 PZQS01000003 PVD35047.1 KK533157 KFP28643.1 KL205917 KFV76655.1 PKK30567.1 KK526502 KFP68314.1 QRBI01000120 RMC06696.1 KB743662 EOA97514.1 KK810732 KFQ35330.1 KK502211 KFP21555.1 AAKN02056864 AAKN02056865 AAKN02056866 AAKN02056867 KK718333 KFO55839.1 KL218306 KFP04203.1 KK750440 KFP41621.1 KK631876 KFV56169.1 AGTP01003739 AGTP01003740 AGTP01003741 AGTP01003742 AGTP01003743 AGTP01003744 KL411326 KFR05628.1 KL456900 KFV09832.1 GBZA01000283 JAG69485.1 AAMC01064616 AAMC01064617 AAMC01064618 AAMC01064619 AAMC01064620 AAMC01064621 AAMC01064622 KL517030 KFO88368.1 KE671461 ERE80238.1 KL355465 KFZ61813.1 LMAW01002756 KQK77446.1 JH167620 EHB02438.1 GEBF01005829 JAN97803.1 KL871155 KGL90914.1 AEMK02000100 DQIR01037948 DQIR01068330 DQIR01073830 DQIR01075520 DQIR01081569 DQIR01088855 DQIR01133935 DQIR01204555 HCZ93423.1 HDA89411.1 KN126200 KFU87496.1 KK444263 KFQ71182.1 JU336956 AFE80709.1 AF071172 AC091304 AC126332 AC135329 AF224243 AF224242 AF224245 AF224244 AF224249 AF224246 AF224247 AF224248 AF224251 AF224250 AF224255 AF224254 AF224252 AF224253 AF224257 AF224256 AF225401 AF225400 AF225404 AF225402 AF225403 AF225407 AF225405 AF225406 AF225409 AF225408 AAD08657.1 AAO27483.1 GFFV01000008 JAV39937.1 AKHW03001628 KYO41478.1 DQIR01081570 DQIR01088852 DQIR01187019 DQIR01311487 HDA37046.1 HDB42496.1 QUSF01000007 RLW07720.1 GAMT01008985 GAMR01001332 GAMQ01000671 GAMP01004912 JAB02876.1 JAB32600.1 JAB41180.1 JAB47843.1 AABR07003539 AABR07003540 AQIB01148333 AQIB01148334 AQIB01148335 AQIB01148336 AQIB01148337 AQIB01148338 AQIB01148339 AQIB01148340 AQIB01148341 AQIB01148342 AMGL01051687 AMGL01051688 AMGL01051689 AMGL01051690 AMGL01051691 AMGL01051692
BABH01002972 NWSH01000054 PCG80163.1 RSAL01000014 RVE53266.1 ODYU01011831 SOQ57796.1 KQ459596 KPI95481.1 KK107167 EZA56377.1 QOIP01000010 RLU17868.1 GL449158 EFN83201.1 GL440194 EFN66182.1 LBMM01000739 KMQ97601.1 KQ971380 KYB24996.1 KK852747 KDR17278.1 ADTU01011956 ADTU01011957 GL888115 EGI67208.1 KQ982490 KYQ55797.1 KZ288445 PBC25641.1 KQ981727 KYN36962.1 KQ978691 KYN29213.1 KQ977408 KYN02952.1 KYB24998.1 KYB24997.1 KQ434831 KZC07832.1 KQ976396 KYM92996.1 GDHC01005102 JAQ13527.1 KB202591 ESO89418.1 LSYS01008075 OPJ69496.1 ABQF01013058 AKCR02000007 PKK30566.1 AADN05000806 ADON01107485 AGTO01010249 PZQS01000003 PVD35047.1 KK533157 KFP28643.1 KL205917 KFV76655.1 PKK30567.1 KK526502 KFP68314.1 QRBI01000120 RMC06696.1 KB743662 EOA97514.1 KK810732 KFQ35330.1 KK502211 KFP21555.1 AAKN02056864 AAKN02056865 AAKN02056866 AAKN02056867 KK718333 KFO55839.1 KL218306 KFP04203.1 KK750440 KFP41621.1 KK631876 KFV56169.1 AGTP01003739 AGTP01003740 AGTP01003741 AGTP01003742 AGTP01003743 AGTP01003744 KL411326 KFR05628.1 KL456900 KFV09832.1 GBZA01000283 JAG69485.1 AAMC01064616 AAMC01064617 AAMC01064618 AAMC01064619 AAMC01064620 AAMC01064621 AAMC01064622 KL517030 KFO88368.1 KE671461 ERE80238.1 KL355465 KFZ61813.1 LMAW01002756 KQK77446.1 JH167620 EHB02438.1 GEBF01005829 JAN97803.1 KL871155 KGL90914.1 AEMK02000100 DQIR01037948 DQIR01068330 DQIR01073830 DQIR01075520 DQIR01081569 DQIR01088855 DQIR01133935 DQIR01204555 HCZ93423.1 HDA89411.1 KN126200 KFU87496.1 KK444263 KFQ71182.1 JU336956 AFE80709.1 AF071172 AC091304 AC126332 AC135329 AF224243 AF224242 AF224245 AF224244 AF224249 AF224246 AF224247 AF224248 AF224251 AF224250 AF224255 AF224254 AF224252 AF224253 AF224257 AF224256 AF225401 AF225400 AF225404 AF225402 AF225403 AF225407 AF225405 AF225406 AF225409 AF225408 AAD08657.1 AAO27483.1 GFFV01000008 JAV39937.1 AKHW03001628 KYO41478.1 DQIR01081570 DQIR01088852 DQIR01187019 DQIR01311487 HDA37046.1 HDB42496.1 QUSF01000007 RLW07720.1 GAMT01008985 GAMR01001332 GAMQ01000671 GAMP01004912 JAB02876.1 JAB32600.1 JAB41180.1 JAB47843.1 AABR07003539 AABR07003540 AQIB01148333 AQIB01148334 AQIB01148335 AQIB01148336 AQIB01148337 AQIB01148338 AQIB01148339 AQIB01148340 AQIB01148341 AQIB01148342 AMGL01051687 AMGL01051688 AMGL01051689 AMGL01051690 AMGL01051691 AMGL01051692
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000053097
UP000279307
+ More
UP000008237 UP000000311 UP000036403 UP000007266 UP000002358 UP000027135 UP000005205 UP000007755 UP000075809 UP000242457 UP000078541 UP000078492 UP000078542 UP000005203 UP000076502 UP000078540 UP000030746 UP000190648 UP000007754 UP000053872 UP000000539 UP000001646 UP000016666 UP000016665 UP000245119 UP000053584 UP000269221 UP000053119 UP000005447 UP000052976 UP000054308 UP000005215 UP000053283 UP000008143 UP000261680 UP000030759 UP000189706 UP000286642 UP000002281 UP000051836 UP000007646 UP000006813 UP000248480 UP000009136 UP000002280 UP000053858 UP000008227 UP000291020 UP000189704 UP000005640 UP000050525 UP000233080 UP000189705 UP000276834 UP000002494 UP000245320 UP000286640 UP000029965 UP000002356
UP000008237 UP000000311 UP000036403 UP000007266 UP000002358 UP000027135 UP000005205 UP000007755 UP000075809 UP000242457 UP000078541 UP000078492 UP000078542 UP000005203 UP000076502 UP000078540 UP000030746 UP000190648 UP000007754 UP000053872 UP000000539 UP000001646 UP000016666 UP000016665 UP000245119 UP000053584 UP000269221 UP000053119 UP000005447 UP000052976 UP000054308 UP000005215 UP000053283 UP000008143 UP000261680 UP000030759 UP000189706 UP000286642 UP000002281 UP000051836 UP000007646 UP000006813 UP000248480 UP000009136 UP000002280 UP000053858 UP000008227 UP000291020 UP000189704 UP000005640 UP000050525 UP000233080 UP000189705 UP000276834 UP000002494 UP000245320 UP000286640 UP000029965 UP000002356
Pfam
Interpro
IPR036400
Cyt_B5-like_heme/steroid_sf
+ More
IPR010606 Mib_Herc2
IPR037252 Mib_Herc2_sf
IPR009091 RCC1/BLIP-II
IPR015940 UBA
IPR004939 APC_su10/DOC_dom
IPR001199 Cyt_B5-like_heme/steroid-bd
IPR008979 Galactose-bd-like_sf
IPR000408 Reg_chr_condens
IPR014722 Rib_L2_dom2
IPR021097 CPH_domain
IPR000569 HECT_dom
IPR035983 Hect_E3_ubiquitin_ligase
IPR006624 Beta-propeller_rpt_TECPR
IPR000433 Znf_ZZ
IPR040847 SH3_15
IPR037976 HERC2_APC10
IPR041987 ZZ_HERC2
IPR016024 ARM-type_fold
IPR013977 GCV_T_C
IPR006076 FAD-dep_OxRdtase
IPR032503 FAO_M
IPR006222 GCV_T_N
IPR027266 TrmE/GcvT_dom1
IPR036188 FAD/NAD-bd_sf
IPR029043 GcvT/YgfZ_C
IPR009060 UBA-like_sf
IPR010606 Mib_Herc2
IPR037252 Mib_Herc2_sf
IPR009091 RCC1/BLIP-II
IPR015940 UBA
IPR004939 APC_su10/DOC_dom
IPR001199 Cyt_B5-like_heme/steroid-bd
IPR008979 Galactose-bd-like_sf
IPR000408 Reg_chr_condens
IPR014722 Rib_L2_dom2
IPR021097 CPH_domain
IPR000569 HECT_dom
IPR035983 Hect_E3_ubiquitin_ligase
IPR006624 Beta-propeller_rpt_TECPR
IPR000433 Znf_ZZ
IPR040847 SH3_15
IPR037976 HERC2_APC10
IPR041987 ZZ_HERC2
IPR016024 ARM-type_fold
IPR013977 GCV_T_C
IPR006076 FAD-dep_OxRdtase
IPR032503 FAO_M
IPR006222 GCV_T_N
IPR027266 TrmE/GcvT_dom1
IPR036188 FAD/NAD-bd_sf
IPR029043 GcvT/YgfZ_C
IPR009060 UBA-like_sf
SUPFAM
ProteinModelPortal
H9JEE6
A0A2A4K8M6
A0A3S2PJI5
A0A2H1WXV6
A0A194PPV9
A0A026WJZ9
+ More
A0A3L8DD68 E2BM75 E2AK78 A0A0J7L4R0 A0A139WAN5 K7IQ42 A0A067R2N5 A0A158NCL1 F4WFF7 A0A151X600 A0A2A3E3G7 A0A195F9S1 A0A195ELX0 A0A195CR34 A0A088AL05 A0A139WAP8 A0A139WAR9 A0A154P7I9 A0A195BYI1 A0A146M433 V3ZE73 A0A1V4JBE1 H0ZJQ4 A0A2I0MLM1 E1BW48 G1KDV1 U3I746 U3JN73 A0A2T7PNV3 A0A091JXI0 A0A093HB52 A0A2I0MLL8 A0A091MCL6 A0A3M0K0M4 A0A3Q2UE20 R0L7U7 A0A091S499 A0A091JP83 A0A286XAB6 H0VHC2 A0A091EHF1 A0A091I9P4 A0A091LFD1 A0A093FPE5 I3M8T9 A0A287CWT3 A0A091VSZ3 A0A093CBX9 A0A0B8RTL9 F7A4S9 A0A384DC01 A0A091HM07 A0A061IGX1 A0A3Q0DBS9 A0A3Q7XI86 A0A094KRD9 A0A3Q2HE57 A0A3Q2IC42 F7AM44 A0A0Q3TAC1 A0A3Q2IA83 G3STX9 G5AZH7 A0A0P6J146 A0A2Y9QW94 E1B782 F7EP78 A0A0A0A8M2 F1RX57 A0A093BN38 A0A091TBI0 H9G1F0 A0A452GTE7 A0A452GSK3 A0A1U7UNG5 O95714 A0A250Y8J0 A0A151NXF6 A0A2K5K3Q5 A0A1U7RFM8 A0A287B1U2 A0A452GSM4 A0A3L8SV73 U3EA95 A0A2K5K3S5 A0A0G2JUC9 A0A452GSJ6 A0A2U4B2F2 A0A3Q7S9L4 A0A0D9R2F7 W5Q2G3
A0A3L8DD68 E2BM75 E2AK78 A0A0J7L4R0 A0A139WAN5 K7IQ42 A0A067R2N5 A0A158NCL1 F4WFF7 A0A151X600 A0A2A3E3G7 A0A195F9S1 A0A195ELX0 A0A195CR34 A0A088AL05 A0A139WAP8 A0A139WAR9 A0A154P7I9 A0A195BYI1 A0A146M433 V3ZE73 A0A1V4JBE1 H0ZJQ4 A0A2I0MLM1 E1BW48 G1KDV1 U3I746 U3JN73 A0A2T7PNV3 A0A091JXI0 A0A093HB52 A0A2I0MLL8 A0A091MCL6 A0A3M0K0M4 A0A3Q2UE20 R0L7U7 A0A091S499 A0A091JP83 A0A286XAB6 H0VHC2 A0A091EHF1 A0A091I9P4 A0A091LFD1 A0A093FPE5 I3M8T9 A0A287CWT3 A0A091VSZ3 A0A093CBX9 A0A0B8RTL9 F7A4S9 A0A384DC01 A0A091HM07 A0A061IGX1 A0A3Q0DBS9 A0A3Q7XI86 A0A094KRD9 A0A3Q2HE57 A0A3Q2IC42 F7AM44 A0A0Q3TAC1 A0A3Q2IA83 G3STX9 G5AZH7 A0A0P6J146 A0A2Y9QW94 E1B782 F7EP78 A0A0A0A8M2 F1RX57 A0A093BN38 A0A091TBI0 H9G1F0 A0A452GTE7 A0A452GSK3 A0A1U7UNG5 O95714 A0A250Y8J0 A0A151NXF6 A0A2K5K3Q5 A0A1U7RFM8 A0A287B1U2 A0A452GSM4 A0A3L8SV73 U3EA95 A0A2K5K3S5 A0A0G2JUC9 A0A452GSJ6 A0A2U4B2F2 A0A3Q7S9L4 A0A0D9R2F7 W5Q2G3
PDB
4L1M
E-value=9.30907e-62,
Score=606
Ontologies
KEGG
GO
Topology
Subcellular location
Cytoplasm
Cytoskeleton
Microtubule organizing center
Centrosome
Centriole
Nucleus
Cytoskeleton
Microtubule organizing center
Centrosome
Centriole
Nucleus
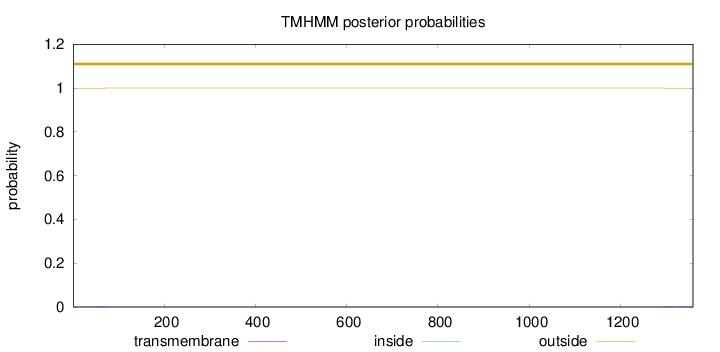
Length:
1359
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03242
Exp number, first 60 AAs:
0.00628
Total prob of N-in:
0.00060
outside
1 - 1359
Population Genetic Test Statistics
Pi
212.086645
Theta
193.255961
Tajima's D
0.873891
CLR
0.419599
CSRT
0.620268986550672
Interpretation
Uncertain
