Gene
KWMTBOMO08762
Pre Gene Modal
BGIBMGA007683
Annotation
death-associated_small_cytoplasmic_leucine-rich_protein_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.26
Sequence
CDS
ATGCGGCCTCCCACGGAATATGACAGCGGTGCGCGCTTGCCCGCCGAAGCCGAGGAGCATGATCCTAGGGCGCCGGCGATGATGATCTGCGCCAGGCTTGCCGGGAGAGCTATCATCAGGGTGGTCGGACGTTGCGAAGATGCCCAAGAGAATAGTCAACTCGATCTATCAGAGTGTCAGCTGATGCAAGTTCCAGATGCAGTATATCACCTAATGAGACACACAGAATTGAAAAGTTGTGATCTCAGTGGGAATGTCATCACTAAAATACCTCCCAAATTTGCAGTCAAGTTTAGTCTGATTACAGATCTCAACCTATCAAACAACCAAATGGCCAGGCTGCCCGATGAGCTATGTACACTTGCTTGTCTACAGCGATTGGACATCTCACATAATACATTCGTAGCTTTACCTCACATAACGTTCCAGTGTCCAAGTCTGCACACATTGCTCGCACATCACAATCAAATTATTGAAGTTGATGTGGACAGATTAGCAAGATCTCAAGCCCTCGAATATGTAGATCTTAGTGACAATCCCATACCTGCACGGACACATGATGACCTGAAACAACTGTCCCGGCCATCAGTAACTTTATCTGATAGACAGAAAGAAGACTGGGAAGAAGATCTCATACTCTAA
Protein
MRPPTEYDSGARLPAEAEEHDPRAPAMMICARLAGRAIIRVVGRCEDAQENSQLDLSECQLMQVPDAVYHLMRHTELKSCDLSGNVITKIPPKFAVKFSLITDLNLSNNQMARLPDELCTLACLQRLDISHNTFVALPHITFQCPSLHTLLAHHNQIIEVDVDRLARSQALEYVDLSDNPIPARTHDDLKQLSRPSVTLSDRQKEDWEEDLIL
Summary
Uniprot
B9VTR9
Q9NGL0
A0A437BS42
A0A212ER35
A0A194QMU5
H9JDT6
+ More
I4DNW4 A0A2A4K891 A0A0L7L8K6 S4PNK3 A0A0L7KVQ4 A0A194PR93 A0A2H1WXX0 A0A0N1IPY2 A0A0T6B1C1 N6TSF7 A0A232F5R5 A0A1B6I5E4 A0A310SQG4 A0A151XE98 A0A1B6GG62 A0A1B6LH32 A0A2A3EH66 A0A195E006 A0A195F4D6 A0A158NV35 E9J2Y3 A0A195BNW4 A0A0M9A8H0 E2C6T1 A0A1Y1NBH7 E2A0X4 A0A195CFG6 A0A026WSE9 A0A1W4X8A4 A0A1B6CLP0 T1H8K3 A0A0A9W046 A0A023F1G8 A0A154PI23 A0A0C9RXR9 A0A0L7R577 A0A088ACE7 V9II26 E0VWX6 F4WS38 A0A146KWA3 A0A2R7W3T4 D6X4G5 A0A1B0GAK9 A0A1B0F933 A0A1J1J6L3 A0A336LW96 D3TQR1 A0A2M4CRT7 A0A1B0GJQ8 A0A1L8DJM7 T1DH26 A0A0C9S0D4 A0A1Q3FHD9 B0WDY2 A0A1Q3FB92 A0A1D2MNE3 A0A0K8W017 A0A182Y6G2 Q17DH1 A0A034VQS6 A0A0A1X7B7 A0A182UVI0 A0A0K8TRY2 A0A182LEQ5 A0A1I8JVQ7 A0A2C9GPX6 Q7QCD5 A0A336LJ76 A0A0J7NQ97 A0A1B0A0Z1 W8B6F4 A0A182INR8 A0A1B0C6Y6 A0A1A9VUM3 A0A084WRI5 A0A182FNV1 A0A1J1J954 A0A023EKM7 A0A034VSC7 A0A034VPS4 A0A0L0BU29 A0A034VMB2 A0A182W0Z4 A0A0M5J1C3 A0A182RC02 A0A226EHM5 A0A034VQT1 A0A1I8NV37 A0A1Q3FHP1 A0A0K8UYA9 A0A182NRK0 A0A182P075
I4DNW4 A0A2A4K891 A0A0L7L8K6 S4PNK3 A0A0L7KVQ4 A0A194PR93 A0A2H1WXX0 A0A0N1IPY2 A0A0T6B1C1 N6TSF7 A0A232F5R5 A0A1B6I5E4 A0A310SQG4 A0A151XE98 A0A1B6GG62 A0A1B6LH32 A0A2A3EH66 A0A195E006 A0A195F4D6 A0A158NV35 E9J2Y3 A0A195BNW4 A0A0M9A8H0 E2C6T1 A0A1Y1NBH7 E2A0X4 A0A195CFG6 A0A026WSE9 A0A1W4X8A4 A0A1B6CLP0 T1H8K3 A0A0A9W046 A0A023F1G8 A0A154PI23 A0A0C9RXR9 A0A0L7R577 A0A088ACE7 V9II26 E0VWX6 F4WS38 A0A146KWA3 A0A2R7W3T4 D6X4G5 A0A1B0GAK9 A0A1B0F933 A0A1J1J6L3 A0A336LW96 D3TQR1 A0A2M4CRT7 A0A1B0GJQ8 A0A1L8DJM7 T1DH26 A0A0C9S0D4 A0A1Q3FHD9 B0WDY2 A0A1Q3FB92 A0A1D2MNE3 A0A0K8W017 A0A182Y6G2 Q17DH1 A0A034VQS6 A0A0A1X7B7 A0A182UVI0 A0A0K8TRY2 A0A182LEQ5 A0A1I8JVQ7 A0A2C9GPX6 Q7QCD5 A0A336LJ76 A0A0J7NQ97 A0A1B0A0Z1 W8B6F4 A0A182INR8 A0A1B0C6Y6 A0A1A9VUM3 A0A084WRI5 A0A182FNV1 A0A1J1J954 A0A023EKM7 A0A034VSC7 A0A034VPS4 A0A0L0BU29 A0A034VMB2 A0A182W0Z4 A0A0M5J1C3 A0A182RC02 A0A226EHM5 A0A034VQT1 A0A1I8NV37 A0A1Q3FHP1 A0A0K8UYA9 A0A182NRK0 A0A182P075
Pubmed
10590172
22118469
26354079
19121390
22651552
26227816
+ More
23622113 23537049 28648823 21347285 21282665 20798317 28004739 24508170 30249741 25401762 26823975 25474469 20566863 21719571 18362917 19820115 20353571 27289101 25244985 17510324 25348373 25830018 26369729 20966253 12364791 14747013 17210077 24495485 24438588 24945155 26108605
23622113 23537049 28648823 21347285 21282665 20798317 28004739 24508170 30249741 25401762 26823975 25474469 20566863 21719571 18362917 19820115 20353571 27289101 25244985 17510324 25348373 25830018 26369729 20966253 12364791 14747013 17210077 24495485 24438588 24945155 26108605
EMBL
FJ602779
ACM24345.1
AF250910
AAF65467.1
RSAL01000014
RVE53264.1
+ More
AGBW02013164 OWR43921.1 KQ461195 KPJ06669.1 BABH01002977 AK403173 BAM19604.1 NWSH01000054 PCG80148.1 JTDY01002316 KOB71675.1 GAIX01000817 JAA91743.1 JTDY01005110 KOB67362.1 KQ459596 KPI95478.1 ODYU01011471 SOQ57224.1 KQ460139 KPJ17516.1 LJIG01016315 KRT80971.1 APGK01053904 KB741238 KB632314 ENN72185.1 ERL92188.1 NNAY01000865 OXU26154.1 GECU01025557 GECU01006005 JAS82149.1 JAT01702.1 KQ760132 OAD61819.1 KQ982254 KYQ58679.1 GECZ01008342 GECZ01004331 JAS61427.1 JAS65438.1 GEBQ01017093 JAT22884.1 KZ288256 PBC30622.1 KQ980031 KYN18194.1 KQ981820 KYN35246.1 ADTU01026906 GL768055 EFZ12788.1 KQ976424 KYM88207.1 KQ435712 KOX79377.1 GL453161 EFN76393.1 GEZM01007292 JAV95261.1 GL435626 EFN73027.1 KQ977827 KYM99460.1 KK107135 QOIP01000006 EZA58014.1 RLU21682.1 GEDC01023000 JAS14298.1 ACPB03001243 GBHO01041792 GBRD01012891 GDHC01018282 GDHC01010662 JAG01812.1 JAG52935.1 JAQ00347.1 JAQ07967.1 GBBI01003345 JAC15367.1 KQ434923 KZC11511.1 GBYB01014015 JAG83782.1 KQ414654 KOC66040.1 JR047029 AEY60307.1 DS235824 EEB17882.1 GL888296 EGI62966.1 GDHC01017866 JAQ00763.1 KK854291 PTY14239.1 KQ971380 EEZ97272.1 CCAG010019543 CVRI01000074 CRL08053.1 UFQS01000021 UFQT01000021 SSW97522.1 SSX17908.1 EZ423763 ADD20039.1 GGFL01003874 MBW68052.1 AJWK01022088 AJWK01022089 GFDF01007540 JAV06544.1 GAMD01002531 JAA99059.1 GBYB01014016 JAG83783.1 GFDL01008069 JAV26976.1 DS231902 EDS45063.1 GFDL01010277 JAV24768.1 LJIJ01000791 ODM94546.1 GDHF01007878 GDHF01000399 JAI44436.1 JAI51915.1 CH477295 EAT44413.1 GAKP01014480 JAC44472.1 GBXI01010352 GBXI01007098 JAD03940.1 JAD07194.1 GDAI01000690 JAI16913.1 APCN01000306 APCN01000307 AAAB01008859 EAA08048.3 SSW97524.1 SSX17910.1 LBMM01002595 KMQ94600.1 GAMC01012378 JAB94177.1 JXJN01027224 ATLV01026091 ATLV01026092 ATLV01026093 KE525406 KFB52829.1 CRL08054.1 GAPW01003896 JAC09702.1 GAKP01014479 JAC44473.1 GAKP01014478 JAC44474.1 JRES01001334 KNC23585.1 GAKP01014476 JAC44476.1 CP012528 ALC49611.1 LNIX01000003 OXA57193.1 GAKP01014475 JAC44477.1 GFDL01007965 JAV27080.1 GDHF01020657 JAI31657.1
AGBW02013164 OWR43921.1 KQ461195 KPJ06669.1 BABH01002977 AK403173 BAM19604.1 NWSH01000054 PCG80148.1 JTDY01002316 KOB71675.1 GAIX01000817 JAA91743.1 JTDY01005110 KOB67362.1 KQ459596 KPI95478.1 ODYU01011471 SOQ57224.1 KQ460139 KPJ17516.1 LJIG01016315 KRT80971.1 APGK01053904 KB741238 KB632314 ENN72185.1 ERL92188.1 NNAY01000865 OXU26154.1 GECU01025557 GECU01006005 JAS82149.1 JAT01702.1 KQ760132 OAD61819.1 KQ982254 KYQ58679.1 GECZ01008342 GECZ01004331 JAS61427.1 JAS65438.1 GEBQ01017093 JAT22884.1 KZ288256 PBC30622.1 KQ980031 KYN18194.1 KQ981820 KYN35246.1 ADTU01026906 GL768055 EFZ12788.1 KQ976424 KYM88207.1 KQ435712 KOX79377.1 GL453161 EFN76393.1 GEZM01007292 JAV95261.1 GL435626 EFN73027.1 KQ977827 KYM99460.1 KK107135 QOIP01000006 EZA58014.1 RLU21682.1 GEDC01023000 JAS14298.1 ACPB03001243 GBHO01041792 GBRD01012891 GDHC01018282 GDHC01010662 JAG01812.1 JAG52935.1 JAQ00347.1 JAQ07967.1 GBBI01003345 JAC15367.1 KQ434923 KZC11511.1 GBYB01014015 JAG83782.1 KQ414654 KOC66040.1 JR047029 AEY60307.1 DS235824 EEB17882.1 GL888296 EGI62966.1 GDHC01017866 JAQ00763.1 KK854291 PTY14239.1 KQ971380 EEZ97272.1 CCAG010019543 CVRI01000074 CRL08053.1 UFQS01000021 UFQT01000021 SSW97522.1 SSX17908.1 EZ423763 ADD20039.1 GGFL01003874 MBW68052.1 AJWK01022088 AJWK01022089 GFDF01007540 JAV06544.1 GAMD01002531 JAA99059.1 GBYB01014016 JAG83783.1 GFDL01008069 JAV26976.1 DS231902 EDS45063.1 GFDL01010277 JAV24768.1 LJIJ01000791 ODM94546.1 GDHF01007878 GDHF01000399 JAI44436.1 JAI51915.1 CH477295 EAT44413.1 GAKP01014480 JAC44472.1 GBXI01010352 GBXI01007098 JAD03940.1 JAD07194.1 GDAI01000690 JAI16913.1 APCN01000306 APCN01000307 AAAB01008859 EAA08048.3 SSW97524.1 SSX17910.1 LBMM01002595 KMQ94600.1 GAMC01012378 JAB94177.1 JXJN01027224 ATLV01026091 ATLV01026092 ATLV01026093 KE525406 KFB52829.1 CRL08054.1 GAPW01003896 JAC09702.1 GAKP01014479 JAC44473.1 GAKP01014478 JAC44474.1 JRES01001334 KNC23585.1 GAKP01014476 JAC44476.1 CP012528 ALC49611.1 LNIX01000003 OXA57193.1 GAKP01014475 JAC44477.1 GFDL01007965 JAV27080.1 GDHF01020657 JAI31657.1
Proteomes
UP000283053
UP000007151
UP000053240
UP000005204
UP000218220
UP000037510
+ More
UP000053268 UP000019118 UP000030742 UP000215335 UP000075809 UP000242457 UP000078492 UP000078541 UP000005205 UP000078540 UP000053105 UP000008237 UP000000311 UP000078542 UP000053097 UP000279307 UP000192223 UP000015103 UP000076502 UP000053825 UP000005203 UP000009046 UP000007755 UP000007266 UP000092444 UP000092443 UP000183832 UP000092461 UP000002320 UP000094527 UP000076408 UP000008820 UP000075903 UP000075882 UP000076407 UP000075840 UP000007062 UP000036403 UP000092445 UP000075880 UP000092460 UP000078200 UP000030765 UP000069272 UP000037069 UP000075920 UP000092553 UP000075900 UP000198287 UP000095300 UP000075884 UP000075885
UP000053268 UP000019118 UP000030742 UP000215335 UP000075809 UP000242457 UP000078492 UP000078541 UP000005205 UP000078540 UP000053105 UP000008237 UP000000311 UP000078542 UP000053097 UP000279307 UP000192223 UP000015103 UP000076502 UP000053825 UP000005203 UP000009046 UP000007755 UP000007266 UP000092444 UP000092443 UP000183832 UP000092461 UP000002320 UP000094527 UP000076408 UP000008820 UP000075903 UP000075882 UP000076407 UP000075840 UP000007062 UP000036403 UP000092445 UP000075880 UP000092460 UP000078200 UP000030765 UP000069272 UP000037069 UP000075920 UP000092553 UP000075900 UP000198287 UP000095300 UP000075884 UP000075885
Interpro
SUPFAM
SSF56112
SSF56112
Gene 3D
ProteinModelPortal
B9VTR9
Q9NGL0
A0A437BS42
A0A212ER35
A0A194QMU5
H9JDT6
+ More
I4DNW4 A0A2A4K891 A0A0L7L8K6 S4PNK3 A0A0L7KVQ4 A0A194PR93 A0A2H1WXX0 A0A0N1IPY2 A0A0T6B1C1 N6TSF7 A0A232F5R5 A0A1B6I5E4 A0A310SQG4 A0A151XE98 A0A1B6GG62 A0A1B6LH32 A0A2A3EH66 A0A195E006 A0A195F4D6 A0A158NV35 E9J2Y3 A0A195BNW4 A0A0M9A8H0 E2C6T1 A0A1Y1NBH7 E2A0X4 A0A195CFG6 A0A026WSE9 A0A1W4X8A4 A0A1B6CLP0 T1H8K3 A0A0A9W046 A0A023F1G8 A0A154PI23 A0A0C9RXR9 A0A0L7R577 A0A088ACE7 V9II26 E0VWX6 F4WS38 A0A146KWA3 A0A2R7W3T4 D6X4G5 A0A1B0GAK9 A0A1B0F933 A0A1J1J6L3 A0A336LW96 D3TQR1 A0A2M4CRT7 A0A1B0GJQ8 A0A1L8DJM7 T1DH26 A0A0C9S0D4 A0A1Q3FHD9 B0WDY2 A0A1Q3FB92 A0A1D2MNE3 A0A0K8W017 A0A182Y6G2 Q17DH1 A0A034VQS6 A0A0A1X7B7 A0A182UVI0 A0A0K8TRY2 A0A182LEQ5 A0A1I8JVQ7 A0A2C9GPX6 Q7QCD5 A0A336LJ76 A0A0J7NQ97 A0A1B0A0Z1 W8B6F4 A0A182INR8 A0A1B0C6Y6 A0A1A9VUM3 A0A084WRI5 A0A182FNV1 A0A1J1J954 A0A023EKM7 A0A034VSC7 A0A034VPS4 A0A0L0BU29 A0A034VMB2 A0A182W0Z4 A0A0M5J1C3 A0A182RC02 A0A226EHM5 A0A034VQT1 A0A1I8NV37 A0A1Q3FHP1 A0A0K8UYA9 A0A182NRK0 A0A182P075
I4DNW4 A0A2A4K891 A0A0L7L8K6 S4PNK3 A0A0L7KVQ4 A0A194PR93 A0A2H1WXX0 A0A0N1IPY2 A0A0T6B1C1 N6TSF7 A0A232F5R5 A0A1B6I5E4 A0A310SQG4 A0A151XE98 A0A1B6GG62 A0A1B6LH32 A0A2A3EH66 A0A195E006 A0A195F4D6 A0A158NV35 E9J2Y3 A0A195BNW4 A0A0M9A8H0 E2C6T1 A0A1Y1NBH7 E2A0X4 A0A195CFG6 A0A026WSE9 A0A1W4X8A4 A0A1B6CLP0 T1H8K3 A0A0A9W046 A0A023F1G8 A0A154PI23 A0A0C9RXR9 A0A0L7R577 A0A088ACE7 V9II26 E0VWX6 F4WS38 A0A146KWA3 A0A2R7W3T4 D6X4G5 A0A1B0GAK9 A0A1B0F933 A0A1J1J6L3 A0A336LW96 D3TQR1 A0A2M4CRT7 A0A1B0GJQ8 A0A1L8DJM7 T1DH26 A0A0C9S0D4 A0A1Q3FHD9 B0WDY2 A0A1Q3FB92 A0A1D2MNE3 A0A0K8W017 A0A182Y6G2 Q17DH1 A0A034VQS6 A0A0A1X7B7 A0A182UVI0 A0A0K8TRY2 A0A182LEQ5 A0A1I8JVQ7 A0A2C9GPX6 Q7QCD5 A0A336LJ76 A0A0J7NQ97 A0A1B0A0Z1 W8B6F4 A0A182INR8 A0A1B0C6Y6 A0A1A9VUM3 A0A084WRI5 A0A182FNV1 A0A1J1J954 A0A023EKM7 A0A034VSC7 A0A034VPS4 A0A0L0BU29 A0A034VMB2 A0A182W0Z4 A0A0M5J1C3 A0A182RC02 A0A226EHM5 A0A034VQT1 A0A1I8NV37 A0A1Q3FHP1 A0A0K8UYA9 A0A182NRK0 A0A182P075
PDB
4R5C
E-value=1.27052e-09,
Score=147
Ontologies
KEGG
GO
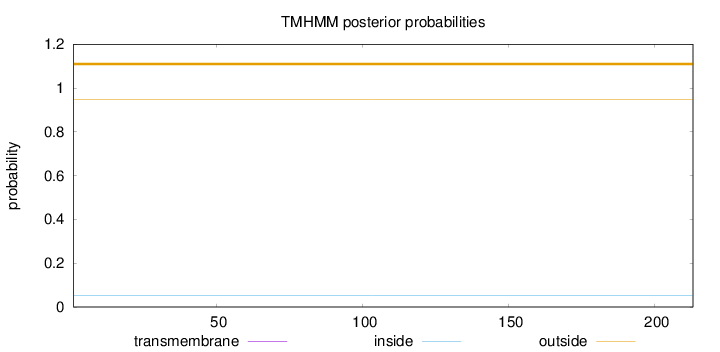
Topology
Length:
213
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00385
Exp number, first 60 AAs:
0.00172
Total prob of N-in:
0.05317
outside
1 - 213
Population Genetic Test Statistics
Pi
200.831234
Theta
177.130064
Tajima's D
-1.783086
CLR
0.897091
CSRT
0.0297485125743713
Interpretation
Uncertain
