Gene
KWMTBOMO08761
Pre Gene Modal
BGIBMGA007682
Annotation
PREDICTED:_PDZ_domain-containing_protein_8_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.163
Sequence
CDS
ATGCCGGACTGGACTTGGATGTTGTTTTCGGCGCTATTCGGAGCTGTGTCTGCGCTAACTGTTCCACTGTTACTTCTCATGTATTACGTACGGCCGTTTGAAAAGAGAACTCAAGAACTGGACCAATTTAACATGCCAGTCACTTTGCCTCTGGAAATTCTGAACGAGCTTACACGTGGAGCCGGCAGCACTCCCGAGGGCGCTCTGAACGCGATGCTGCAGTTCATATTCCAACAAAGCACTGCTGATACTCGCTGGTTACGACGCCGTATGGCTACCGAACTAGCTGAGTTGCTTGCTAAAACATCTTCAGGAAAGATATTTGAATCTCTAACGCTACGTTCCTTGGAAGTTGGCACTGAGTTTCCTAGTATCAACAATCTTCACCTACTTAGCGGCGAATTGGAGGAAGATGGGGCAAGACTCAGGTCCTTGGATCTAAAATTCGATCTTGCCTACGACGGAAACTTCAGGATTGACGTAGACGCTGTGATGCTGCTCGGCAAGATTGCTAATATATCGATTACTGTTGAAAGTCTAAGCGGCAATGTACGACTCATGTTTCGACGCACTCCGTATACCCATTGGGCGCTTGCTTTTGAAGCGCCGCCAAAGTTGGAGTTGAAAGTCCGAGGTCGATTCCAAGGGCGGCAATTAAAGCCAAGACTGGCATCGCTGGTCGCGGCCCATATCAGGAGGGCAGTCGCACAACGGCACACCCTTCCCAATTATAAGATAAGATACAAACCATTCTTCCCAAAATCTGAGCCGGGTAGTGGAGAAGCTGATGACGGTCCCACCCCTCAAGGTCGACTGACGGTACGACTTGTCGAAGTTACCCGATTGCAGTGGGCAGGAGCGAATGGGACAGTAAGGGCTGCTTTAGCTGTGGATTCACATGCATGGGTATCCCTACGTCGTGGTGTTTTAGTGTTGGATGTAGTGTTGCCAGCAGTCTCCGGTCCGCTTGGCCTCGTTTGCTGCCAGGGTGTCGGCGTGGTGCTGGTAGATACAGTAGTGCCAATGTCGCCGGCATATCGCGCCGATTTACGACGAGACGATGTTGTACTGGCCATCGATAATAAGCCTATTAATAATGTCGCTCAGGTTGGCAAACTCCTTCGTCTGGTAGAAAGACGTGCTGTCACCGTGCGAGTTCGACGAGGCGGCAGTGGAAACTCAGCGCGCCGTGAAGACAACGCCGTCGAACCGAAACGGATCAAACCTAACTTTTCATTCAGAAGAGTTTCCGAAGTAATGGAGGGAGTTAGACTGCAGGGTATTAGAGCTGCGGCTTCTAAGAGTAATTTACAGGAATCCGAATCACCGGTTAAAGCTTCTGTTGAATCACACAACGAGACTGACGCACAATCCAAACCAAAAACAGAACCGGTTCGTCAACTTTCCAAGTCAAATGACCAGCCTTCACCTTCAGCCAATTTCGGTCTTCGTAAGCGCCGTTGTTCCGTGGAAAATAAATCTTCCGATAGTTCTGCGGGCAGCACACCACCGGCTTCTGGAGCCAGTACACCCTTACGACAGTCCACCGCTGTGAACAAAACCGTCAAACACCCCGAAATACCTATTATTAGAACTGATTCCTCCGAATGTAAAGTTACGGAGAGAGTTGAAACACTTTTAGATGATGATGAATTATTCGAAACAGGCGGCCCCAGACAATGCGAACATGTTGAGATATGCCAAATGTTTTTGACTAAAAGCATCCAGCTCAAGCCCATCATACCGTTTGATGAAGAAACCGAATTCAAATTGAACGAGAATCATAAATATTTAAATATAAACGTATGGGCAACGGTAACAGGAGAGAAAGATGAAGTTCTGTTGGGATACTTGAATATTCCACTAGCCGCGTTATTGAGCGAATGCCAGGACTCGACAGTTCCACATCATATGAAACGATATCAGTTTTTACCTCCCGATGTCGATATTAAATTTAGCAAAAGCCACAAACTGGCTTCACATGCAGGCTTCGAGCCATGTCTATGTTTTGGAGACGCTCTGGTTTGGTGGGAATGGGATGGGGCAGCTACTCGCAGCGCTCCTCGTCCTCCTGTGAAAGACTCAGCGCCTCCTCGGCCCCCCAAATTGACCAGAGCTGTTCACGATTTTGTACGAACTCACTTCCACAGATCTACTCAATGCGATTTTTGCAATAAAAAGATTTGGTTGAAAGACGCAATGCAATGCCGTAGTTGTGGTCTTTGTTGCCATAAAAAATGCGTTACAAAGTGTCAAGAGTCTTCGCCGTGCGTACCTAAACAAGACTTAGGCTTGAGCTGTCCTCCTCCTGAACTGATTATGACAGAAGCTGACTCGGTTCCCGATCCTGACTCCGACGAAGAACCCGATAGAAAAGATTCACTTGAAGTTCATGAAGAAGGCGGCGGCGCTATGAGCGCGCTGGTGGCGGGGCTACGGCGTGCTGGCTCTGCACACTCGCTCTCGGCTCCCAAAACGTCTGCTTCTTCCAGGTCCTTACCACCCTCACCAAGCCATACCCCCAGGAAGCAGTCGCTGGAGTCGGTGGGCAACCCGTTCGGCGCGTGTGGCGTGACGTGGGCACAGCGGGGCGACGTGAGCGCGGTGCTGGCGCCTGCGCTGCGCAGCGCTCTGCCGCCGGACGCGCTGCTCGCGGCCGCCAGGGACGCGGCGCCTGATCTCGCCGCCAGCCTCTCCGCTCAACAGATACACGATATGTTACTGGCAGTTCGTGAGGAATTATCAGCAGAAGATGCAAGTGACCCAGACGGGGCTGGTGGCGAAGGACGGGCATACGCACTAACAGCGCTGATGCTGCACTGCTGTGCCGCCGCACAACTAGCGCAGCAACGTGCTGACGCCGCACCTGATAACACCTAA
Protein
MPDWTWMLFSALFGAVSALTVPLLLLMYYVRPFEKRTQELDQFNMPVTLPLEILNELTRGAGSTPEGALNAMLQFIFQQSTADTRWLRRRMATELAELLAKTSSGKIFESLTLRSLEVGTEFPSINNLHLLSGELEEDGARLRSLDLKFDLAYDGNFRIDVDAVMLLGKIANISITVESLSGNVRLMFRRTPYTHWALAFEAPPKLELKVRGRFQGRQLKPRLASLVAAHIRRAVAQRHTLPNYKIRYKPFFPKSEPGSGEADDGPTPQGRLTVRLVEVTRLQWAGANGTVRAALAVDSHAWVSLRRGVLVLDVVLPAVSGPLGLVCCQGVGVVLVDTVVPMSPAYRADLRRDDVVLAIDNKPINNVAQVGKLLRLVERRAVTVRVRRGGSGNSARREDNAVEPKRIKPNFSFRRVSEVMEGVRLQGIRAAASKSNLQESESPVKASVESHNETDAQSKPKTEPVRQLSKSNDQPSPSANFGLRKRRCSVENKSSDSSAGSTPPASGASTPLRQSTAVNKTVKHPEIPIIRTDSSECKVTERVETLLDDDELFETGGPRQCEHVEICQMFLTKSIQLKPIIPFDEETEFKLNENHKYLNINVWATVTGEKDEVLLGYLNIPLAALLSECQDSTVPHHMKRYQFLPPDVDIKFSKSHKLASHAGFEPCLCFGDALVWWEWDGAATRSAPRPPVKDSAPPRPPKLTRAVHDFVRTHFHRSTQCDFCNKKIWLKDAMQCRSCGLCCHKKCVTKCQESSPCVPKQDLGLSCPPPELIMTEADSVPDPDSDEEPDRKDSLEVHEEGGGAMSALVAGLRRAGSAHSLSAPKTSASSRSLPPSPSHTPRKQSLESVGNPFGACGVTWAQRGDVSAVLAPALRSALPPDALLAAARDAAPDLAASLSAQQIHDMLLAVREELSAEDASDPDGAGGEGRAYALTALMLHCCAAAQLAQQRADAAPDNT
Summary
Uniprot
EMBL
Proteomes
PRIDE
Interpro
SUPFAM
SSF50156
SSF50156
CDD
ProteinModelPortal
PDB
4L9M
E-value=0.00142931,
Score=103
Ontologies
GO
PANTHER
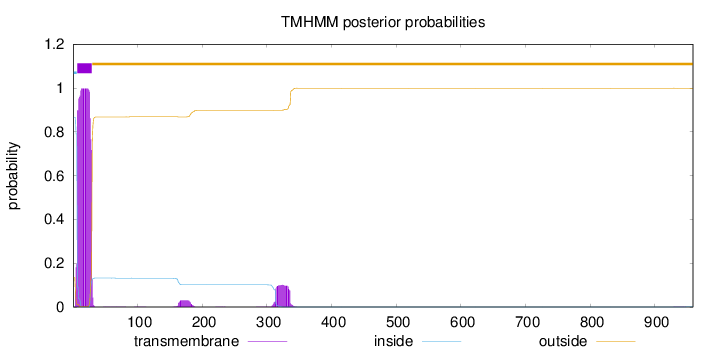
Topology
Length:
959
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
25.5780899999999
Exp number, first 60 AAs:
22.5865
Total prob of N-in:
0.86557
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 29
outside
30 - 959
Population Genetic Test Statistics
Pi
233.343043
Theta
173.18716
Tajima's D
1.1999
CLR
0.396237
CSRT
0.709764511774411
Interpretation
Uncertain
