Gene
KWMTBOMO08760 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007681
Annotation
PREDICTED:_phosphoglycerate_kinase_[Amyelois_transitella]
Full name
Phosphoglycerate kinase
Location in the cell
Cytoplasmic Reliability : 4.126
Sequence
CDS
ATGGCTTTAAACAAATTAAGTATTGATGCACTTAACTTAACCGGCAAAAGAGTTCTAATGCGTGTGGATTTCAATGTACCGCTTAAAGAAGGAGTGATTACAAACAACCAGCGCATTGTAGCTGCCTTAGATTCCGTTAAATATGCTTTGGATAAAGGCGCTAAATCAGTTGTACTTATGTCGCATTTGGGTAGACCGGATGGCCAGGTAAATCTCAAGTACACCCTCAAACCAGTCGCTGAAGAGCTAAAAAAATTGCTGAACAAGGATGTGACATTTTTGAATGACTGCATAGGCCCGGAGGTTGAAACAGCATGTGCTAATCCTTCAGCTGGTTCAATTATCCTACTGGAGAATCTTCGCTTCCACATTGAAGAGGAAGGCAAAGGTGTAGATGCTTCAGGTGCTAAAGTGAAGGCTGATCCAGAAAAAGTTAAAGCCTTTAGGGCAAGCTTGAGAAAACTTGGTGATGTCTATATTAATGATGCTTTTGGTACTGCACATAGAGCGCACAGTTCAATGGTGGGAGAAGGTTTTGAGCAGAGGGCCAGTGGTTTCTTACTAAAGAAAGAGCTTCAATACTTTGCCAAAGCTTTACATGAACCTGAGAGGCCCTTCCTGGCTATCCTTGGTGGAGCTAAAGTAGCAGACAAAATCTTACTTATAGAGAACCTGCTTGATAAAGTAAATGAAATGATAATTGGAGGGGGTATGGCATACACATTCTTGAAGGAAACCAAAGGAATGCCAATTGGTAATTCCCTGTATGACGCTGAGGGAGCTAAAATTGTGACAAAACTATTGGAAAAAGCTGAGAAAAACAATGTTAAAGTTCACCTGCCTGTTGATTTTGTGACAGCTGATAAATTTGATGAAAATGCCTCGGTGGGAGAAGCTAATGTTGAAACCGGAATTCCTGATGGATGGATGGGACTCGATGTTGGACCGAAATCACGAGAGCTCTTCGCAGATCCTATTGCCAGAGCTAAAGTTATTGTGTGGAATGGACCTGCTGGCGTGTTTGAGTTTGAGAAATTCGCAGGTGGAACCCGAGCCATCATGGATGGTGTCGTAAAGGCTACATCCAATGGAACCGTCACAATCATTGGTGGTGGTGATACTGCAACGTGTTGCGCCAAGTGGGGAACAGAGGACAAGGTCTCGCACGTCTCTACTGGCGGTGGAGCTTCTCTGGAGTTACTCGAAGGAAAAGTGCTTCCCGGTGTTGCCGCTCTTTCAGATGCATAA
Protein
MALNKLSIDALNLTGKRVLMRVDFNVPLKEGVITNNQRIVAALDSVKYALDKGAKSVVLMSHLGRPDGQVNLKYTLKPVAEELKKLLNKDVTFLNDCIGPEVETACANPSAGSIILLENLRFHIEEEGKGVDASGAKVKADPEKVKAFRASLRKLGDVYINDAFGTAHRAHSSMVGEGFEQRASGFLLKKELQYFAKALHEPERPFLAILGGAKVADKILLIENLLDKVNEMIIGGGMAYTFLKETKGMPIGNSLYDAEGAKIVTKLLEKAEKNNVKVHLPVDFVTADKFDENASVGEANVETGIPDGWMGLDVGPKSRELFADPIARAKVIVWNGPAGVFEFEKFAGGTRAIMDGVVKATSNGTVTIIGGGDTATCCAKWGTEDKVSHVSTGGGASLELLEGKVLPGVAALSDA
Summary
Catalytic Activity
3-phospho-D-glycerate + ATP = 3-phospho-D-glyceroyl phosphate + ADP
Subunit
Monomer.
Similarity
Belongs to the phosphoglycerate kinase family.
Keywords
ATP-binding
Complete proteome
Cytoplasm
Glycolysis
Kinase
Nucleotide-binding
Reference proteome
Transferase
Feature
chain Phosphoglycerate kinase
Uniprot
H9JDT4
A0A2A4K7E2
A0A437BS31
A0A194QMB2
S4PSF1
A0A0K2CTV7
+ More
A0A212ER03 A0A194PWG7 I4DJT8 V5GXP0 A0A1S3D5F9 A0A1Y1KSN7 A0A0A9YUG7 D2A5H8 A0A0M8ZPV2 A0A1Y1KTC3 A0A0L7RD63 J3JZG7 A0A195FVQ9 A0A151ITJ5 E9IUG2 A0A151XA97 E2AJ35 A0A3L8DHW4 A0A0T6AU40 A0A026WTQ9 A0A411G6S5 F4WLH1 A0A2H8TL24 A0A0K8TTK7 K7IM64 A0A0C5QH08 A0A151IDB9 J9K4W4 A0A087ZNJ8 Q5XUA9 A0A0J7L696 A0A0P4WMP5 A0A1W4WCB9 A0A1B6FQ82 A0A069DU88 A0A154PA08 A0A195AV49 V9IIT6 A0A158NNB5 A0A224XD72 A0A232EYI8 A0A1I9WLE6 A0A1S3D3Y6 A0A1W4WND0 A0A2S2PXU7 A0A423SCY3 A0A067R8S1 A0A1B0CUP3 A0A1L8E1W2 A0A1I8NZ87 A0A1L8EGZ9 A0A1B6CFY2 A0A0L0C8V6 T1P9Y2 A0A1L8E220 T1I4E2 A0A087TPE1 A0A0V0G5S4 A0A0P4VNT3 A0A034WAC5 B4JDD7 A0A0C9RNL4 A0A1V9XJU4 A0A170Z687 B4KLD0 B4NWT7 A0A1J1IPJ5 A0A1W4UTF3 W8C486 B4LUU2 B3NA50 A0A0K8V856 E2BGV9 A5XCH6 A5XCI5 M9PCE0 Q01604 A5XCI1 B3N1A0 A0A3B0K898 B4I2P3 Q29KW9 B4GSP3 R4WDR4 A0A1W7RA08 A0A2P2HZI5 A0A1S3HH77 B4N0G7 A0A443SA22 E9FRQ6 A0A336LY55 A0A1Q3FM24 A0A336N2R9 B0WPW1
A0A212ER03 A0A194PWG7 I4DJT8 V5GXP0 A0A1S3D5F9 A0A1Y1KSN7 A0A0A9YUG7 D2A5H8 A0A0M8ZPV2 A0A1Y1KTC3 A0A0L7RD63 J3JZG7 A0A195FVQ9 A0A151ITJ5 E9IUG2 A0A151XA97 E2AJ35 A0A3L8DHW4 A0A0T6AU40 A0A026WTQ9 A0A411G6S5 F4WLH1 A0A2H8TL24 A0A0K8TTK7 K7IM64 A0A0C5QH08 A0A151IDB9 J9K4W4 A0A087ZNJ8 Q5XUA9 A0A0J7L696 A0A0P4WMP5 A0A1W4WCB9 A0A1B6FQ82 A0A069DU88 A0A154PA08 A0A195AV49 V9IIT6 A0A158NNB5 A0A224XD72 A0A232EYI8 A0A1I9WLE6 A0A1S3D3Y6 A0A1W4WND0 A0A2S2PXU7 A0A423SCY3 A0A067R8S1 A0A1B0CUP3 A0A1L8E1W2 A0A1I8NZ87 A0A1L8EGZ9 A0A1B6CFY2 A0A0L0C8V6 T1P9Y2 A0A1L8E220 T1I4E2 A0A087TPE1 A0A0V0G5S4 A0A0P4VNT3 A0A034WAC5 B4JDD7 A0A0C9RNL4 A0A1V9XJU4 A0A170Z687 B4KLD0 B4NWT7 A0A1J1IPJ5 A0A1W4UTF3 W8C486 B4LUU2 B3NA50 A0A0K8V856 E2BGV9 A5XCH6 A5XCI5 M9PCE0 Q01604 A5XCI1 B3N1A0 A0A3B0K898 B4I2P3 Q29KW9 B4GSP3 R4WDR4 A0A1W7RA08 A0A2P2HZI5 A0A1S3HH77 B4N0G7 A0A443SA22 E9FRQ6 A0A336LY55 A0A1Q3FM24 A0A336N2R9 B0WPW1
EC Number
2.7.2.3
Pubmed
19121390
26354079
23622113
26263512
22118469
22651552
+ More
28004739 25401762 18362917 19820115 22516182 21282665 20798317 30249741 24508170 21719571 26369729 20075255 25702953 26334808 21347285 28648823 27538518 24845553 26108605 25315136 27129103 25348373 17994087 28327890 17550304 24495485 17379620 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 1465095 15632085 23691247 21292972
28004739 25401762 18362917 19820115 22516182 21282665 20798317 30249741 24508170 21719571 26369729 20075255 25702953 26334808 21347285 28648823 27538518 24845553 26108605 25315136 27129103 25348373 17994087 28327890 17550304 24495485 17379620 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 1465095 15632085 23691247 21292972
EMBL
BABH01002981
BABH01002982
NWSH01000054
PCG80151.1
RSAL01000014
RVE53262.1
+ More
KQ461195 KPJ06667.1 GAIX01013898 JAA78662.1 KT218673 ODYU01001750 ALA09393.1 SOQ38379.1 AGBW02013164 OWR43923.1 KQ459596 KPI95475.1 AK401556 BAM18178.1 GALX01003348 JAB65118.1 GEZM01077016 JAV63461.1 GBHO01006982 JAG36622.1 KQ971345 EFA05374.1 KQ438551 KOX67203.1 GEZM01077017 JAV63460.1 KQ414614 KOC68798.1 BT128649 AEE63606.1 KQ981208 KYN44725.1 KQ981037 KYN10165.1 GL765974 EFZ15795.1 KQ982351 KYQ57292.1 GL439932 EFN66567.1 QOIP01000008 RLU19941.1 LJIG01022804 KRT78626.1 KK107109 EZA59046.1 MH365801 QBB01610.1 GL888208 EGI64907.1 GFXV01002183 MBW13988.1 GDAI01000117 JAI17486.1 KM609490 AJQ30114.1 KQ977978 KYM98370.1 ABLF02022778 AY737545 AAU84938.1 LBMM01000586 KMQ98019.1 GDRN01007624 JAI67940.1 GECZ01017407 JAS52362.1 GBGD01001622 JAC87267.1 KQ434856 KZC08671.1 KQ976738 KYM75849.1 JR047391 AEY60401.1 ADTU01021203 GFTR01006020 JAW10406.1 NNAY01001594 OXU23484.1 KU932330 APA33966.1 GGMS01000987 MBY70190.1 QCYY01003863 ROT62024.1 KK852809 KDR15993.1 AJWK01029481 AJWK01029482 GFDF01001361 JAV12723.1 GFDG01000851 JAV17948.1 GEDC01024960 JAS12338.1 JRES01000905 JRES01000841 KNC27364.1 KNC27829.1 KA645581 AFP60210.1 GFDF01001360 JAV12724.1 ACPB03010950 KK116158 KFM66980.1 GECL01003405 JAP02719.1 GDKW01000231 JAI56364.1 GAKP01008239 JAC50713.1 CH916368 EDW03307.1 GBYB01008671 JAG78438.1 MNPL01009444 OQR73726.1 GEMB01002575 JAS00614.1 CH933807 EDW12811.1 CM000157 EDW87429.1 CVRI01000055 CRL01468.1 GAMC01009726 JAB96829.1 CH940649 EDW64269.1 CH954177 EDV57513.1 GDHF01017278 JAI35036.1 GL448228 EFN85091.1 DQ863977 DQ863978 DQ863979 DQ863980 DQ863981 DQ863983 DQ863984 DQ863985 DQ863987 DQ863988 CM000361 CM002910 ABH06612.1 ABH06613.1 ABH06614.1 ABH06615.1 ABH06616.1 ABH06618.1 ABH06619.1 ABH06620.1 ABH06622.1 ABH06623.1 EDX03510.1 KMY87723.1 DQ863986 ABH06621.1 AE014134 AGB92495.1 Z14029 DQ863952 DQ863953 DQ863954 DQ863955 DQ863956 DQ863957 DQ863958 DQ863959 DQ863960 DQ863961 DQ863962 DQ863963 DQ863964 DQ863965 DQ863966 DQ863967 DQ863968 DQ863969 DQ863970 DQ863971 DQ863972 DQ863973 DQ863974 DQ863975 DQ863976 BT023910 DQ863982 ABH06617.1 CH902652 EDV33621.1 OUUW01000006 SPP81856.1 CH480820 EDW54038.1 CH379061 EAL33054.1 CH479189 EDW25402.1 AK417908 BAN21123.1 GFAH01000433 JAV47956.1 IACF01001318 LAB67030.1 CH963920 EDW77580.1 NCKV01004948 RWS24383.1 GL732523 EFX90443.1 UFQS01000219 UFQT01000219 SSX01541.1 SSX21921.1 GFDL01006375 JAV28670.1 UFQT01003592 SSX35127.1 DS232030 EDS32549.1
KQ461195 KPJ06667.1 GAIX01013898 JAA78662.1 KT218673 ODYU01001750 ALA09393.1 SOQ38379.1 AGBW02013164 OWR43923.1 KQ459596 KPI95475.1 AK401556 BAM18178.1 GALX01003348 JAB65118.1 GEZM01077016 JAV63461.1 GBHO01006982 JAG36622.1 KQ971345 EFA05374.1 KQ438551 KOX67203.1 GEZM01077017 JAV63460.1 KQ414614 KOC68798.1 BT128649 AEE63606.1 KQ981208 KYN44725.1 KQ981037 KYN10165.1 GL765974 EFZ15795.1 KQ982351 KYQ57292.1 GL439932 EFN66567.1 QOIP01000008 RLU19941.1 LJIG01022804 KRT78626.1 KK107109 EZA59046.1 MH365801 QBB01610.1 GL888208 EGI64907.1 GFXV01002183 MBW13988.1 GDAI01000117 JAI17486.1 KM609490 AJQ30114.1 KQ977978 KYM98370.1 ABLF02022778 AY737545 AAU84938.1 LBMM01000586 KMQ98019.1 GDRN01007624 JAI67940.1 GECZ01017407 JAS52362.1 GBGD01001622 JAC87267.1 KQ434856 KZC08671.1 KQ976738 KYM75849.1 JR047391 AEY60401.1 ADTU01021203 GFTR01006020 JAW10406.1 NNAY01001594 OXU23484.1 KU932330 APA33966.1 GGMS01000987 MBY70190.1 QCYY01003863 ROT62024.1 KK852809 KDR15993.1 AJWK01029481 AJWK01029482 GFDF01001361 JAV12723.1 GFDG01000851 JAV17948.1 GEDC01024960 JAS12338.1 JRES01000905 JRES01000841 KNC27364.1 KNC27829.1 KA645581 AFP60210.1 GFDF01001360 JAV12724.1 ACPB03010950 KK116158 KFM66980.1 GECL01003405 JAP02719.1 GDKW01000231 JAI56364.1 GAKP01008239 JAC50713.1 CH916368 EDW03307.1 GBYB01008671 JAG78438.1 MNPL01009444 OQR73726.1 GEMB01002575 JAS00614.1 CH933807 EDW12811.1 CM000157 EDW87429.1 CVRI01000055 CRL01468.1 GAMC01009726 JAB96829.1 CH940649 EDW64269.1 CH954177 EDV57513.1 GDHF01017278 JAI35036.1 GL448228 EFN85091.1 DQ863977 DQ863978 DQ863979 DQ863980 DQ863981 DQ863983 DQ863984 DQ863985 DQ863987 DQ863988 CM000361 CM002910 ABH06612.1 ABH06613.1 ABH06614.1 ABH06615.1 ABH06616.1 ABH06618.1 ABH06619.1 ABH06620.1 ABH06622.1 ABH06623.1 EDX03510.1 KMY87723.1 DQ863986 ABH06621.1 AE014134 AGB92495.1 Z14029 DQ863952 DQ863953 DQ863954 DQ863955 DQ863956 DQ863957 DQ863958 DQ863959 DQ863960 DQ863961 DQ863962 DQ863963 DQ863964 DQ863965 DQ863966 DQ863967 DQ863968 DQ863969 DQ863970 DQ863971 DQ863972 DQ863973 DQ863974 DQ863975 DQ863976 BT023910 DQ863982 ABH06617.1 CH902652 EDV33621.1 OUUW01000006 SPP81856.1 CH480820 EDW54038.1 CH379061 EAL33054.1 CH479189 EDW25402.1 AK417908 BAN21123.1 GFAH01000433 JAV47956.1 IACF01001318 LAB67030.1 CH963920 EDW77580.1 NCKV01004948 RWS24383.1 GL732523 EFX90443.1 UFQS01000219 UFQT01000219 SSX01541.1 SSX21921.1 GFDL01006375 JAV28670.1 UFQT01003592 SSX35127.1 DS232030 EDS32549.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000007151
UP000053268
+ More
UP000079169 UP000007266 UP000053105 UP000053825 UP000078541 UP000078492 UP000075809 UP000000311 UP000279307 UP000053097 UP000007755 UP000002358 UP000078542 UP000007819 UP000005203 UP000036403 UP000192223 UP000076502 UP000078540 UP000005205 UP000215335 UP000283509 UP000027135 UP000092461 UP000095300 UP000037069 UP000095301 UP000015103 UP000054359 UP000001070 UP000192247 UP000009192 UP000002282 UP000183832 UP000192221 UP000008792 UP000008711 UP000008237 UP000000304 UP000000803 UP000007801 UP000268350 UP000001292 UP000001819 UP000008744 UP000085678 UP000007798 UP000288716 UP000000305 UP000002320
UP000079169 UP000007266 UP000053105 UP000053825 UP000078541 UP000078492 UP000075809 UP000000311 UP000279307 UP000053097 UP000007755 UP000002358 UP000078542 UP000007819 UP000005203 UP000036403 UP000192223 UP000076502 UP000078540 UP000005205 UP000215335 UP000283509 UP000027135 UP000092461 UP000095300 UP000037069 UP000095301 UP000015103 UP000054359 UP000001070 UP000192247 UP000009192 UP000002282 UP000183832 UP000192221 UP000008792 UP000008711 UP000008237 UP000000304 UP000000803 UP000007801 UP000268350 UP000001292 UP000001819 UP000008744 UP000085678 UP000007798 UP000288716 UP000000305 UP000002320
PRIDE
Interpro
IPR001576
Phosphoglycerate_kinase
+ More
IPR015824 Phosphoglycerate_kinase_N
IPR036043 Phosphoglycerate_kinase_sf
IPR015911 Phosphoglycerate_kinase_CS
IPR041591 OCRE
IPR000467 G_patch_dom
IPR035979 RBD_domain_sf
IPR036443 Znf_RanBP2_sf
IPR013087 Znf_C2H2_type
IPR001876 Znf_RanBP2
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR015824 Phosphoglycerate_kinase_N
IPR036043 Phosphoglycerate_kinase_sf
IPR015911 Phosphoglycerate_kinase_CS
IPR041591 OCRE
IPR000467 G_patch_dom
IPR035979 RBD_domain_sf
IPR036443 Znf_RanBP2_sf
IPR013087 Znf_C2H2_type
IPR001876 Znf_RanBP2
IPR012677 Nucleotide-bd_a/b_plait_sf
Gene 3D
ProteinModelPortal
H9JDT4
A0A2A4K7E2
A0A437BS31
A0A194QMB2
S4PSF1
A0A0K2CTV7
+ More
A0A212ER03 A0A194PWG7 I4DJT8 V5GXP0 A0A1S3D5F9 A0A1Y1KSN7 A0A0A9YUG7 D2A5H8 A0A0M8ZPV2 A0A1Y1KTC3 A0A0L7RD63 J3JZG7 A0A195FVQ9 A0A151ITJ5 E9IUG2 A0A151XA97 E2AJ35 A0A3L8DHW4 A0A0T6AU40 A0A026WTQ9 A0A411G6S5 F4WLH1 A0A2H8TL24 A0A0K8TTK7 K7IM64 A0A0C5QH08 A0A151IDB9 J9K4W4 A0A087ZNJ8 Q5XUA9 A0A0J7L696 A0A0P4WMP5 A0A1W4WCB9 A0A1B6FQ82 A0A069DU88 A0A154PA08 A0A195AV49 V9IIT6 A0A158NNB5 A0A224XD72 A0A232EYI8 A0A1I9WLE6 A0A1S3D3Y6 A0A1W4WND0 A0A2S2PXU7 A0A423SCY3 A0A067R8S1 A0A1B0CUP3 A0A1L8E1W2 A0A1I8NZ87 A0A1L8EGZ9 A0A1B6CFY2 A0A0L0C8V6 T1P9Y2 A0A1L8E220 T1I4E2 A0A087TPE1 A0A0V0G5S4 A0A0P4VNT3 A0A034WAC5 B4JDD7 A0A0C9RNL4 A0A1V9XJU4 A0A170Z687 B4KLD0 B4NWT7 A0A1J1IPJ5 A0A1W4UTF3 W8C486 B4LUU2 B3NA50 A0A0K8V856 E2BGV9 A5XCH6 A5XCI5 M9PCE0 Q01604 A5XCI1 B3N1A0 A0A3B0K898 B4I2P3 Q29KW9 B4GSP3 R4WDR4 A0A1W7RA08 A0A2P2HZI5 A0A1S3HH77 B4N0G7 A0A443SA22 E9FRQ6 A0A336LY55 A0A1Q3FM24 A0A336N2R9 B0WPW1
A0A212ER03 A0A194PWG7 I4DJT8 V5GXP0 A0A1S3D5F9 A0A1Y1KSN7 A0A0A9YUG7 D2A5H8 A0A0M8ZPV2 A0A1Y1KTC3 A0A0L7RD63 J3JZG7 A0A195FVQ9 A0A151ITJ5 E9IUG2 A0A151XA97 E2AJ35 A0A3L8DHW4 A0A0T6AU40 A0A026WTQ9 A0A411G6S5 F4WLH1 A0A2H8TL24 A0A0K8TTK7 K7IM64 A0A0C5QH08 A0A151IDB9 J9K4W4 A0A087ZNJ8 Q5XUA9 A0A0J7L696 A0A0P4WMP5 A0A1W4WCB9 A0A1B6FQ82 A0A069DU88 A0A154PA08 A0A195AV49 V9IIT6 A0A158NNB5 A0A224XD72 A0A232EYI8 A0A1I9WLE6 A0A1S3D3Y6 A0A1W4WND0 A0A2S2PXU7 A0A423SCY3 A0A067R8S1 A0A1B0CUP3 A0A1L8E1W2 A0A1I8NZ87 A0A1L8EGZ9 A0A1B6CFY2 A0A0L0C8V6 T1P9Y2 A0A1L8E220 T1I4E2 A0A087TPE1 A0A0V0G5S4 A0A0P4VNT3 A0A034WAC5 B4JDD7 A0A0C9RNL4 A0A1V9XJU4 A0A170Z687 B4KLD0 B4NWT7 A0A1J1IPJ5 A0A1W4UTF3 W8C486 B4LUU2 B3NA50 A0A0K8V856 E2BGV9 A5XCH6 A5XCI5 M9PCE0 Q01604 A5XCI1 B3N1A0 A0A3B0K898 B4I2P3 Q29KW9 B4GSP3 R4WDR4 A0A1W7RA08 A0A2P2HZI5 A0A1S3HH77 B4N0G7 A0A443SA22 E9FRQ6 A0A336LY55 A0A1Q3FM24 A0A336N2R9 B0WPW1
PDB
2PAA
E-value=1.40849e-174,
Score=1574
Ontologies
PATHWAY
GO
PANTHER
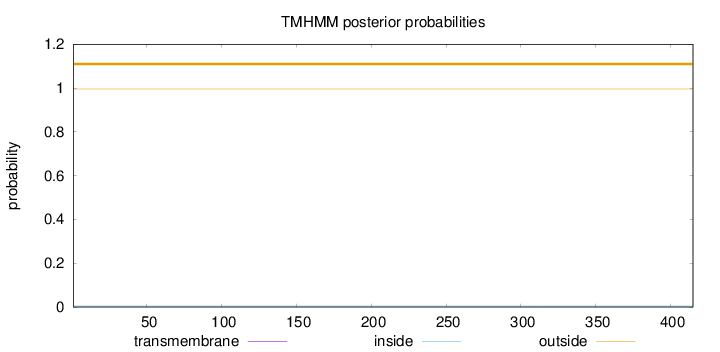
Topology
Subcellular location
Cytoplasm
Length:
415
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00339000000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00451
outside
1 - 415
Population Genetic Test Statistics
Pi
154.29116
Theta
166.249333
Tajima's D
0.479752
CLR
0.142851
CSRT
0.511974401279936
Interpretation
Uncertain
