Gene
KWMTBOMO08754 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007899
Annotation
peritrophin-1-like_precursor_[Papilio_xuthus]
Location in the cell
Extracellular Reliability : 3.658
Sequence
CDS
ATGAGAGTGTTCATCGTATTGACGGCAGTCGCTGCAATTGCAAGCGCCCAATTCAAGTGCCCCGCTAAAGACGGCCAGTATGAAGACGACAGACAATGCGACAAGTTCTTCGAATGTGTCGATGGAGTCGCCACAACCAAGCTCTGCCCGGACGGTCTTGTGTTCGACCCTACTATCAGGAAGATCAACAAATGCGACCAGCCCTTCAACGTAGACTGCGGCGATAGGACTGAACTCCAGCCCCCCAAGCCCAACTCACAATGTCCCCGCCGCAATGGATTCTTTGCCCATCCCGACCCCTCTGTGTGCAACATCTTCTACAACTGCATCGAAGGTGAAGCCACCGAGGTCAAGTGTACCGCCGGCCTTCACTTCGACGAGTACTCCGGTACCTGCGTCTGGCCCGATTCCGCCGGAAGACAAGGATGTAACGAACAACAAAAAAAAACAAAGGACGGCTTCGAGTGCCCCAAAGAGCAGCTGGTTGATGCCCAAGGACAGATCGTCGCTCACCCCAAGTTCCCCCACCCCAACGACTGCCAACGTTTCTACGTATGCCTGAACGGTGTCGAGCCCCGTGACCTCGGCTGCCAAGTCGGAGAGGTTTATAACGAAGAGAGCCAGAAATGCGATGCGCCTGAGAACGTTCGCGGATGCGAGGACTGGTACAAGGACTCTGAGGAAGCCGCACCAGGACCCAAATCGAGATCTTAA
Protein
MRVFIVLTAVAAIASAQFKCPAKDGQYEDDRQCDKFFECVDGVATTKLCPDGLVFDPTIRKINKCDQPFNVDCGDRTELQPPKPNSQCPRRNGFFAHPDPSVCNIFYNCIEGEATEVKCTAGLHFDEYSGTCVWPDSAGRQGCNEQQKKTKDGFECPKEQLVDAQGQIVAHPKFPHPNDCQRFYVCLNGVEPRDLGCQVGEVYNEESQKCDAPENVRGCEDWYKDSEEAAPGPKSRS
Summary
Uniprot
H9JEF2
I4DIS0
A0A194PWG1
I4DM10
A0A2A4JQJ5
A0A2W1BJB4
+ More
A0A2H1W7M0 A0A194QMB1 A0A212EYJ0 A0A3S2LGE1 G9D4W5 G9D4X2 A0A0L7L0G2 A0A182W553 A0A182J939 G9D4W4 A0A1I8JUT6 D1MAI4 G9D4W7 G9D4W9 T1DJ78 A0A084VJF9 G9D4X6 Q16MN2 G9D4W8 A0A1S4GAI5 A0A1Y9GLX8 G9D4W6 G9D4X7 G9D4X4 G9D4X5 G9D4X1 G9D4X0 A0A023EKL3 A0A1Q3FPB3 G9D4X3 Q7QHZ7 B0W0L0 W5JJF9 A0A1L8E3C7 A0A182XNG4 A0A1L8E3M5 A0A182GD32 A0A182U9E4 A0A182QG74 A0A0F6PN76 A0A3Q8HPY9 A0A182SG84 A0A1B0CEA6 A0A088AGM5 J3JY26 U4UAM8 A0A158NER9 A0A3L8E3V2 E2B783 A0A182N0U3 A0A310SNY5 A0A067R898 A0A195FMZ9 A0A2P8Z085 A0A0C9RTG6 A0A1Y1KJ01 F4WEF0 E2A164 A0A1B0G8D3 A0A1A9Z5U4 A0A1A9XXT9 A0A1B0APQ2 B4L7X1 A0A1A9W224 A0A1A9VBM3 A0A0K8TNN7 A0A1B0DAP5 A0A182PKY3 A0A182M4T1 A0A146LKW4 A0A0A9Y133 A0A0L0BSS6 A0A2K8JM15 A0A1I8PCV1 A0A1W4UQZ3 B4PY11 Q9VR79 A0A224XR67 A0A0Q9WQC5 B3NWG8 A0A1B6E516 A0A161MJN8 B4JKA0 A0A182K5K0 A0A232EME8 A0A3B0J857 B4M9Y4 A0A0M4ETU9 Q29GW4 B4GV26 A0A2J7PZW8 T1P866 A0A023FA42 A0A0P4VRI5
A0A2H1W7M0 A0A194QMB1 A0A212EYJ0 A0A3S2LGE1 G9D4W5 G9D4X2 A0A0L7L0G2 A0A182W553 A0A182J939 G9D4W4 A0A1I8JUT6 D1MAI4 G9D4W7 G9D4W9 T1DJ78 A0A084VJF9 G9D4X6 Q16MN2 G9D4W8 A0A1S4GAI5 A0A1Y9GLX8 G9D4W6 G9D4X7 G9D4X4 G9D4X5 G9D4X1 G9D4X0 A0A023EKL3 A0A1Q3FPB3 G9D4X3 Q7QHZ7 B0W0L0 W5JJF9 A0A1L8E3C7 A0A182XNG4 A0A1L8E3M5 A0A182GD32 A0A182U9E4 A0A182QG74 A0A0F6PN76 A0A3Q8HPY9 A0A182SG84 A0A1B0CEA6 A0A088AGM5 J3JY26 U4UAM8 A0A158NER9 A0A3L8E3V2 E2B783 A0A182N0U3 A0A310SNY5 A0A067R898 A0A195FMZ9 A0A2P8Z085 A0A0C9RTG6 A0A1Y1KJ01 F4WEF0 E2A164 A0A1B0G8D3 A0A1A9Z5U4 A0A1A9XXT9 A0A1B0APQ2 B4L7X1 A0A1A9W224 A0A1A9VBM3 A0A0K8TNN7 A0A1B0DAP5 A0A182PKY3 A0A182M4T1 A0A146LKW4 A0A0A9Y133 A0A0L0BSS6 A0A2K8JM15 A0A1I8PCV1 A0A1W4UQZ3 B4PY11 Q9VR79 A0A224XR67 A0A0Q9WQC5 B3NWG8 A0A1B6E516 A0A161MJN8 B4JKA0 A0A182K5K0 A0A232EME8 A0A3B0J857 B4M9Y4 A0A0M4ETU9 Q29GW4 B4GV26 A0A2J7PZW8 T1P866 A0A023FA42 A0A0P4VRI5
Pubmed
19121390
22651552
26354079
28756777
22118469
21967427
+ More
26227816 18362917 20144715 19820115 24438588 17510324 12364791 24945155 20920257 23761445 26483478 22516182 23537049 21347285 30249741 20798317 24845553 29403074 28004739 21719571 17994087 26369729 26823975 25401762 26108605 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28648823 15632085 25315136 25474469 27129103
26227816 18362917 20144715 19820115 24438588 17510324 12364791 24945155 20920257 23761445 26483478 22516182 23537049 21347285 30249741 20798317 24845553 29403074 28004739 21719571 17994087 26369729 26823975 25401762 26108605 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28648823 15632085 25315136 25474469 27129103
EMBL
BABH01003008
AK401188
BAM17810.1
KQ459596
KPI95470.1
AK402328
+ More
BAM18950.1 NWSH01000845 PCG73874.1 KZ150017 PZC74958.1 ODYU01006835 SOQ49047.1 KQ461195 KPJ06662.1 AGBW02011497 OWR46555.1 RSAL01000014 RVE53257.1 JF777368 AEU11713.1 JF777375 AEU11720.1 JTDY01003940 KOB68801.1 JF777367 AEU11712.1 GU128092 KQ971379 ACY95475.1 EEZ97329.1 JF777370 AEU11715.1 JF777372 AEU11717.1 GAMD01001601 JAA99989.1 ATLV01013557 KE524867 KFB38103.1 JF777379 AEU11724.1 CH477855 EAT35608.1 JF777371 AEU11716.1 AAAB01008811 APCN01004770 JF777369 AEU11714.1 JF777380 AEU11725.1 JF777377 AEU11722.1 JF777378 AEU11723.1 JF777374 AEU11719.1 JF777373 AEU11718.1 GAPW01003911 JAC09687.1 GFDL01005606 JAV29439.1 JF777376 AEU11721.1 EAA04933.5 DS231818 EDS41312.1 ADMH02000984 ETN64512.1 GFDF01000856 JAV13228.1 GFDF01000855 JAV13229.1 JXUM01055320 KQ561861 KXJ77304.1 AXCN02002174 KM099158 AJZ68819.1 MG601672 AYA49997.1 AJWK01008709 AJWK01008710 APGK01031880 APGK01031881 APGK01031882 APGK01031883 BT128150 KB740829 AEE63111.1 ENN78823.1 KB632205 ERL90117.1 ADTU01013619 ADTU01013620 QOIP01000001 RLU27143.1 GL446149 EFN88444.1 KQ759804 OAD62822.1 KK852869 KDR14646.1 KQ981490 KYN41289.1 PYGN01000257 PSN49911.1 GBYB01010786 JAG80553.1 GEZM01082209 JAV61403.1 GL888103 EGI67437.1 GL435707 EFN72831.1 CCAG010000243 JXJN01001544 JXJN01001545 CH933814 EDW05546.1 GDAI01001867 JAI15736.1 AJVK01004584 AXCM01000244 GDHC01009916 JAQ08713.1 GBHO01016842 JAG26762.1 JRES01001419 KNC23071.1 KY031115 ATU82866.1 CM000162 EDX02983.1 AE014298 BT001512 AAF50927.2 AAN71267.1 AFH07490.1 GFTR01004118 JAW12308.1 CH940655 KRF82549.1 CH954180 EDV46788.1 GEDC01004272 JAS33026.1 GEMB01000885 GEMB01000884 JAS02256.1 CH916370 EDW00003.1 NNAY01003378 OXU19517.1 OUUW01000003 SPP78494.1 EDW66043.1 CP012528 ALC50027.1 CH379064 EAL31995.2 CH479192 EDW26563.1 NEVH01020330 PNF21872.1 KA644777 AFP59406.1 GBBI01000919 JAC17793.1 GDKW01003864 JAI52731.1
BAM18950.1 NWSH01000845 PCG73874.1 KZ150017 PZC74958.1 ODYU01006835 SOQ49047.1 KQ461195 KPJ06662.1 AGBW02011497 OWR46555.1 RSAL01000014 RVE53257.1 JF777368 AEU11713.1 JF777375 AEU11720.1 JTDY01003940 KOB68801.1 JF777367 AEU11712.1 GU128092 KQ971379 ACY95475.1 EEZ97329.1 JF777370 AEU11715.1 JF777372 AEU11717.1 GAMD01001601 JAA99989.1 ATLV01013557 KE524867 KFB38103.1 JF777379 AEU11724.1 CH477855 EAT35608.1 JF777371 AEU11716.1 AAAB01008811 APCN01004770 JF777369 AEU11714.1 JF777380 AEU11725.1 JF777377 AEU11722.1 JF777378 AEU11723.1 JF777374 AEU11719.1 JF777373 AEU11718.1 GAPW01003911 JAC09687.1 GFDL01005606 JAV29439.1 JF777376 AEU11721.1 EAA04933.5 DS231818 EDS41312.1 ADMH02000984 ETN64512.1 GFDF01000856 JAV13228.1 GFDF01000855 JAV13229.1 JXUM01055320 KQ561861 KXJ77304.1 AXCN02002174 KM099158 AJZ68819.1 MG601672 AYA49997.1 AJWK01008709 AJWK01008710 APGK01031880 APGK01031881 APGK01031882 APGK01031883 BT128150 KB740829 AEE63111.1 ENN78823.1 KB632205 ERL90117.1 ADTU01013619 ADTU01013620 QOIP01000001 RLU27143.1 GL446149 EFN88444.1 KQ759804 OAD62822.1 KK852869 KDR14646.1 KQ981490 KYN41289.1 PYGN01000257 PSN49911.1 GBYB01010786 JAG80553.1 GEZM01082209 JAV61403.1 GL888103 EGI67437.1 GL435707 EFN72831.1 CCAG010000243 JXJN01001544 JXJN01001545 CH933814 EDW05546.1 GDAI01001867 JAI15736.1 AJVK01004584 AXCM01000244 GDHC01009916 JAQ08713.1 GBHO01016842 JAG26762.1 JRES01001419 KNC23071.1 KY031115 ATU82866.1 CM000162 EDX02983.1 AE014298 BT001512 AAF50927.2 AAN71267.1 AFH07490.1 GFTR01004118 JAW12308.1 CH940655 KRF82549.1 CH954180 EDV46788.1 GEDC01004272 JAS33026.1 GEMB01000885 GEMB01000884 JAS02256.1 CH916370 EDW00003.1 NNAY01003378 OXU19517.1 OUUW01000003 SPP78494.1 EDW66043.1 CP012528 ALC50027.1 CH379064 EAL31995.2 CH479192 EDW26563.1 NEVH01020330 PNF21872.1 KA644777 AFP59406.1 GBBI01000919 JAC17793.1 GDKW01003864 JAI52731.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000053240
UP000007151
UP000283053
+ More
UP000037510 UP000075920 UP000075880 UP000075900 UP000007266 UP000030765 UP000008820 UP000075840 UP000007062 UP000002320 UP000000673 UP000076407 UP000069940 UP000249989 UP000075902 UP000075886 UP000075901 UP000092461 UP000005203 UP000019118 UP000030742 UP000005205 UP000279307 UP000008237 UP000075884 UP000027135 UP000078541 UP000245037 UP000007755 UP000000311 UP000092444 UP000092445 UP000092443 UP000092460 UP000009192 UP000091820 UP000078200 UP000092462 UP000075885 UP000075883 UP000037069 UP000095300 UP000192221 UP000002282 UP000000803 UP000008792 UP000008711 UP000001070 UP000075881 UP000215335 UP000268350 UP000092553 UP000001819 UP000008744 UP000235965 UP000095301
UP000037510 UP000075920 UP000075880 UP000075900 UP000007266 UP000030765 UP000008820 UP000075840 UP000007062 UP000002320 UP000000673 UP000076407 UP000069940 UP000249989 UP000075902 UP000075886 UP000075901 UP000092461 UP000005203 UP000019118 UP000030742 UP000005205 UP000279307 UP000008237 UP000075884 UP000027135 UP000078541 UP000245037 UP000007755 UP000000311 UP000092444 UP000092445 UP000092443 UP000092460 UP000009192 UP000091820 UP000078200 UP000092462 UP000075885 UP000075883 UP000037069 UP000095300 UP000192221 UP000002282 UP000000803 UP000008792 UP000008711 UP000001070 UP000075881 UP000215335 UP000268350 UP000092553 UP000001819 UP000008744 UP000235965 UP000095301
Pfam
PF01607 CBM_14
SUPFAM
SSF57625
SSF57625
ProteinModelPortal
H9JEF2
I4DIS0
A0A194PWG1
I4DM10
A0A2A4JQJ5
A0A2W1BJB4
+ More
A0A2H1W7M0 A0A194QMB1 A0A212EYJ0 A0A3S2LGE1 G9D4W5 G9D4X2 A0A0L7L0G2 A0A182W553 A0A182J939 G9D4W4 A0A1I8JUT6 D1MAI4 G9D4W7 G9D4W9 T1DJ78 A0A084VJF9 G9D4X6 Q16MN2 G9D4W8 A0A1S4GAI5 A0A1Y9GLX8 G9D4W6 G9D4X7 G9D4X4 G9D4X5 G9D4X1 G9D4X0 A0A023EKL3 A0A1Q3FPB3 G9D4X3 Q7QHZ7 B0W0L0 W5JJF9 A0A1L8E3C7 A0A182XNG4 A0A1L8E3M5 A0A182GD32 A0A182U9E4 A0A182QG74 A0A0F6PN76 A0A3Q8HPY9 A0A182SG84 A0A1B0CEA6 A0A088AGM5 J3JY26 U4UAM8 A0A158NER9 A0A3L8E3V2 E2B783 A0A182N0U3 A0A310SNY5 A0A067R898 A0A195FMZ9 A0A2P8Z085 A0A0C9RTG6 A0A1Y1KJ01 F4WEF0 E2A164 A0A1B0G8D3 A0A1A9Z5U4 A0A1A9XXT9 A0A1B0APQ2 B4L7X1 A0A1A9W224 A0A1A9VBM3 A0A0K8TNN7 A0A1B0DAP5 A0A182PKY3 A0A182M4T1 A0A146LKW4 A0A0A9Y133 A0A0L0BSS6 A0A2K8JM15 A0A1I8PCV1 A0A1W4UQZ3 B4PY11 Q9VR79 A0A224XR67 A0A0Q9WQC5 B3NWG8 A0A1B6E516 A0A161MJN8 B4JKA0 A0A182K5K0 A0A232EME8 A0A3B0J857 B4M9Y4 A0A0M4ETU9 Q29GW4 B4GV26 A0A2J7PZW8 T1P866 A0A023FA42 A0A0P4VRI5
A0A2H1W7M0 A0A194QMB1 A0A212EYJ0 A0A3S2LGE1 G9D4W5 G9D4X2 A0A0L7L0G2 A0A182W553 A0A182J939 G9D4W4 A0A1I8JUT6 D1MAI4 G9D4W7 G9D4W9 T1DJ78 A0A084VJF9 G9D4X6 Q16MN2 G9D4W8 A0A1S4GAI5 A0A1Y9GLX8 G9D4W6 G9D4X7 G9D4X4 G9D4X5 G9D4X1 G9D4X0 A0A023EKL3 A0A1Q3FPB3 G9D4X3 Q7QHZ7 B0W0L0 W5JJF9 A0A1L8E3C7 A0A182XNG4 A0A1L8E3M5 A0A182GD32 A0A182U9E4 A0A182QG74 A0A0F6PN76 A0A3Q8HPY9 A0A182SG84 A0A1B0CEA6 A0A088AGM5 J3JY26 U4UAM8 A0A158NER9 A0A3L8E3V2 E2B783 A0A182N0U3 A0A310SNY5 A0A067R898 A0A195FMZ9 A0A2P8Z085 A0A0C9RTG6 A0A1Y1KJ01 F4WEF0 E2A164 A0A1B0G8D3 A0A1A9Z5U4 A0A1A9XXT9 A0A1B0APQ2 B4L7X1 A0A1A9W224 A0A1A9VBM3 A0A0K8TNN7 A0A1B0DAP5 A0A182PKY3 A0A182M4T1 A0A146LKW4 A0A0A9Y133 A0A0L0BSS6 A0A2K8JM15 A0A1I8PCV1 A0A1W4UQZ3 B4PY11 Q9VR79 A0A224XR67 A0A0Q9WQC5 B3NWG8 A0A1B6E516 A0A161MJN8 B4JKA0 A0A182K5K0 A0A232EME8 A0A3B0J857 B4M9Y4 A0A0M4ETU9 Q29GW4 B4GV26 A0A2J7PZW8 T1P866 A0A023FA42 A0A0P4VRI5
PDB
6G9E
E-value=1.34878e-08,
Score=139
Ontologies
GO
Topology
SignalP
Position: 1 - 16,
Likelihood: 0.999154
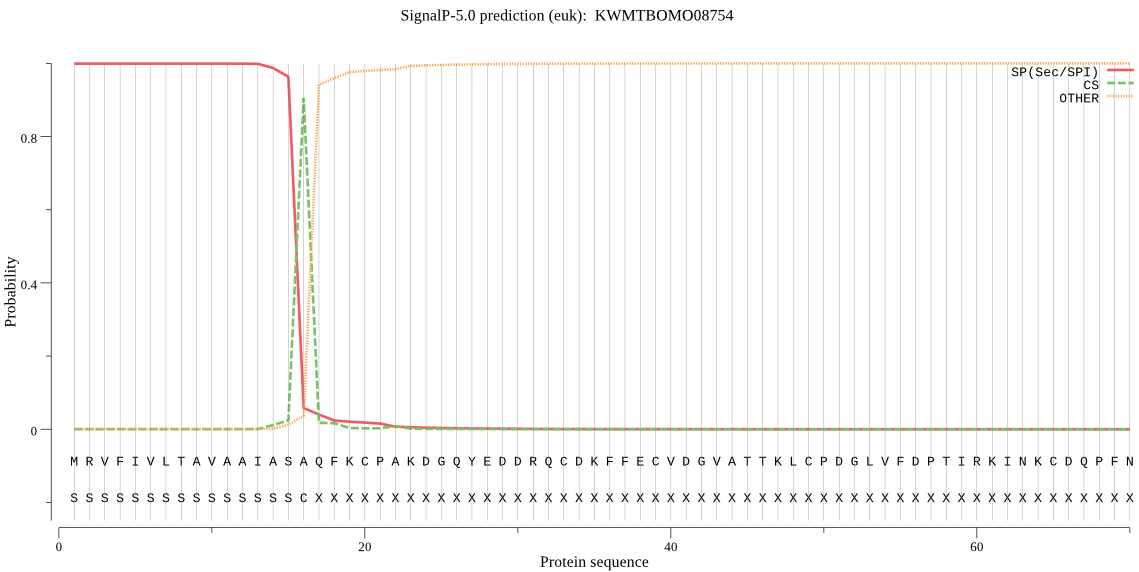
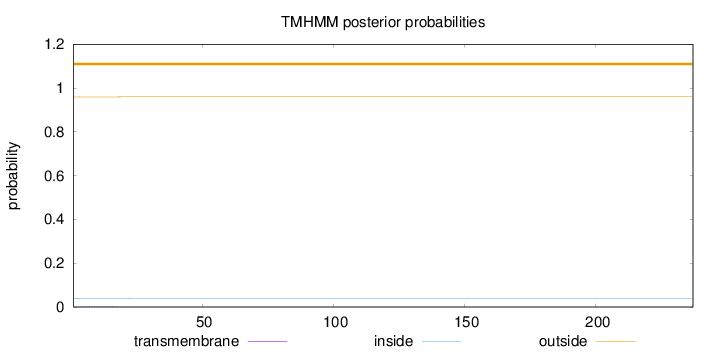
Length:
237
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04168
Exp number, first 60 AAs:
0.04168
Total prob of N-in:
0.04044
outside
1 - 237
Population Genetic Test Statistics
Pi
219.532
Theta
190.377969
Tajima's D
0.019917
CLR
0.093516
CSRT
0.373131343432828
Interpretation
Uncertain
