Gene
KWMTBOMO08753 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007900
Annotation
chondroitin_proteoglycan-2-like_precursor_[Papilio_xuthus]
Location in the cell
Extracellular Reliability : 2.903
Sequence
CDS
ATGAAAAGTTTAATAGTACTAGGGTTGGCTGTTTGTGGATTAGTTTCAGGCCAAGAATTCAAATGCCCGGACAAGAGCGGCTTCTACCCGGACCCGTATCAATGTGACCTTTATTACAAATGCAGCAGAGGAGATGCAGAAGAGAAGCTGTGTCCTGATGGTCTCGTGTTCTCAGATGAAAACCCCAACAAAGAACACTGTGATATCCCTTCTAACGTCGACTGCGGCGACAGAAAGGAGCTACAAGAACCTAAACCATCGAAGGGCTGCCCTCGTCAAAACGGATACTTTAAACATCCTGACCCACAGGCCTGCGATAAATTCCACTACTGCGCTGATGGTATTCCCAACGAGCTTCCGTGTCCTCCAGGACTGTACTTTGACGAAGAGACCTCCAACTGCGACTGGAAAGAAGTAGTCAACAGGCAATGCGACCAAATCACAAAAGATGTTTTGGATGATGGTTTCACCTGTCCCGACGGCGAGGTCATGGGCCCCAACGGCCGCTCCTTACCCCACCCCACTTTCCCCCACCCCGAAGACTGCCAGAAGTTCTATATCTGCCGTAACGGAGTCCAACCGCAAAAGGGAAGCTGTCCATCCGGGAAGGTCTACAATGAGGATACGTTCATGTGCGACGATCCCGAAAAAGTTGTTGGATGTGAAAACTACTACGACGGACAGCCACTTGACAAGAACAAGCTTCCCAAGAAGGCGTAA
Protein
MKSLIVLGLAVCGLVSGQEFKCPDKSGFYPDPYQCDLYYKCSRGDAEEKLCPDGLVFSDENPNKEHCDIPSNVDCGDRKELQEPKPSKGCPRQNGYFKHPDPQACDKFHYCADGIPNELPCPPGLYFDEETSNCDWKEVVNRQCDQITKDVLDDGFTCPDGEVMGPNGRSLPHPTFPHPEDCQKFYICRNGVQPQKGSCPSGKVYNEDTFMCDDPEKVVGCENYYDGQPLDKNKLPKKA
Summary
Uniprot
H9JEF3
I4DJ44
A0A3S2NZP3
A0A194QSZ5
A0A2H1W7G7
A0A212EYI4
+ More
A0A2A4JP85 A0A2W1BNU5 A0A069DPZ6 A0A023F984 A0A2P8Z0B8 A0A2S1ZS45 A0A067QYQ5 A0A1B6HUP9 A0A1B6D180 A0A1Y1NL60 A0A0A9WAZ2 D1MAI5 A0A146LAW5 A0A2J7PZW4 R4WJR8 A0A0F6PN07 A0A1W4XCC5 A0A0A9W8D0 E2B782 F4WEF1 A0A158NER8 A0A2S2Q042 A0A2S2NVZ4 A0A0T6AUY5 A0A154PD68 A0A1S3DGK4 J9JZY1 A0A195FLJ7 R4G5Z9 A0A0L7QP93 A0A195DZL8 A0A3L8E3L9 K7IMV2 A0A232EME2 A0A195AVM1 A0A0J7KJS7 E9IQV8 A0A0C9QWL8 A0A151XIU6 A0A195CR50 A0A026WG30 T1I1T4 A0A0N0BK39 A0A1D2MYL9 A0A087ZT03 A0A1Q3FPB3 B0W0L0 A0A1S4FMS0 Q16VF5 Q178G5 I4DIS0 A0A194PWG1 A0A2H1W7M0 A0A182V3S0 I4DM10 A0A1W7R8Z4 A0A1Y1KJ01 A0A084VJG1 J9K501 A0A1Y9GLC3 A0A2W1BJB4 Q7PFC5 A0A084VJF9 A0A2A4JQJ5 B0W0L2 A0A023EKL3 A0A2S2NBZ9 A0A1S4GAI5 A0A1Y9GLX8 A0A067R898 A0A2K8JM15 A0A1Q3FPL7 A0A2P8Z085 A0A1B6M0M6 A0A2H8TR84 Q7QHZ7 D1MAI4 Q16MN2 A0A1B6L5Z3 A0A1B6E3P5 A0A182J939 A0A224XR67 H9JEF2 T1DJ78 A0A1I8JUT6 A0A182W553 A0A2S1ZS47 A0A1I8JUR0 A0A1B6FE13 A0A182W552 A0A158NER9 A0A1B6E516 A0A182SH98
A0A2A4JP85 A0A2W1BNU5 A0A069DPZ6 A0A023F984 A0A2P8Z0B8 A0A2S1ZS45 A0A067QYQ5 A0A1B6HUP9 A0A1B6D180 A0A1Y1NL60 A0A0A9WAZ2 D1MAI5 A0A146LAW5 A0A2J7PZW4 R4WJR8 A0A0F6PN07 A0A1W4XCC5 A0A0A9W8D0 E2B782 F4WEF1 A0A158NER8 A0A2S2Q042 A0A2S2NVZ4 A0A0T6AUY5 A0A154PD68 A0A1S3DGK4 J9JZY1 A0A195FLJ7 R4G5Z9 A0A0L7QP93 A0A195DZL8 A0A3L8E3L9 K7IMV2 A0A232EME2 A0A195AVM1 A0A0J7KJS7 E9IQV8 A0A0C9QWL8 A0A151XIU6 A0A195CR50 A0A026WG30 T1I1T4 A0A0N0BK39 A0A1D2MYL9 A0A087ZT03 A0A1Q3FPB3 B0W0L0 A0A1S4FMS0 Q16VF5 Q178G5 I4DIS0 A0A194PWG1 A0A2H1W7M0 A0A182V3S0 I4DM10 A0A1W7R8Z4 A0A1Y1KJ01 A0A084VJG1 J9K501 A0A1Y9GLC3 A0A2W1BJB4 Q7PFC5 A0A084VJF9 A0A2A4JQJ5 B0W0L2 A0A023EKL3 A0A2S2NBZ9 A0A1S4GAI5 A0A1Y9GLX8 A0A067R898 A0A2K8JM15 A0A1Q3FPL7 A0A2P8Z085 A0A1B6M0M6 A0A2H8TR84 Q7QHZ7 D1MAI4 Q16MN2 A0A1B6L5Z3 A0A1B6E3P5 A0A182J939 A0A224XR67 H9JEF2 T1DJ78 A0A1I8JUT6 A0A182W553 A0A2S1ZS47 A0A1I8JUR0 A0A1B6FE13 A0A182W552 A0A158NER9 A0A1B6E516 A0A182SH98
Pubmed
EMBL
BABH01003009
AK401312
KQ459596
BAM17934.1
KPI95469.1
RSAL01000014
+ More
RVE53256.1 KQ461195 KPJ06661.1 ODYU01006835 SOQ49045.1 AGBW02011497 OWR46556.1 NWSH01000845 PCG73875.1 KZ150017 PZC74957.1 GBGD01002741 JAC86148.1 GBBI01001153 JAC17559.1 PYGN01000257 PSN49920.1 MF942760 AWK28283.1 KK852869 KDR14645.1 GECU01029308 JAS78398.1 GEDC01017850 JAS19448.1 GEZM01003873 JAV96507.1 GBHO01039941 JAG03663.1 GU128093 KQ971379 ACY95476.1 EEZ97330.1 GDHC01014293 JAQ04336.1 NEVH01020330 PNF21871.1 AK417835 BAN21050.1 KM099159 AJZ68820.1 GBHO01039943 GDHC01004434 JAG03661.1 JAQ14195.1 GL446149 EFN88443.1 GL888103 EGI67438.1 ADTU01013618 GGMS01001841 MBY71044.1 GGMR01008731 MBY21350.1 LJIG01022743 KRT78947.1 KQ434867 KZC09150.1 ABLF02036175 KQ981490 KYN41288.1 GAHY01000075 JAA77435.1 KQ414821 KOC60450.1 KQ979999 KYN18318.1 QOIP01000001 RLU27142.1 NNAY01003378 OXU19516.1 KQ976736 KYM76107.1 LBMM01006569 KMQ90501.1 GL764974 EFZ17045.1 GBYB01000105 JAG69872.1 KQ982080 KYQ60235.1 KQ977381 KYN03181.1 KK107260 EZA53994.1 ACPB03005154 KQ435709 KOX80018.1 LJIJ01000399 ODM97884.1 GFDL01005606 JAV29439.1 DS231818 EDS41312.1 CH477594 EAT38548.1 CH477365 EAT42533.1 AK401188 BAM17810.1 KPI95470.1 SOQ49047.1 AK402328 BAM18950.1 GEHC01000070 JAV47575.1 GEZM01082209 JAV61403.1 ATLV01013558 KE524867 KFB38105.1 APCN01004769 APCN01004770 PZC74958.1 AAAB01008811 EAA45375.2 ATLV01013557 KFB38103.1 PCG73874.1 EDS41314.1 GAPW01003911 JAC09687.1 GGMR01001853 MBY14472.1 KDR14646.1 KY031115 ATU82866.1 GFDL01005593 JAV29452.1 PSN49911.1 GEBQ01010488 JAT29489.1 GFXV01004909 MBW16714.1 EAA04933.5 GU128092 ACY95475.1 EEZ97329.1 CH477855 EAT35608.1 GEBQ01020891 JAT19086.1 GEDC01004779 JAS32519.1 GFTR01004118 JAW12308.1 BABH01003008 GAMD01001601 JAA99989.1 MF942759 AWK28282.1 GECZ01021342 JAS48427.1 ADTU01013619 ADTU01013620 GEDC01004272 JAS33026.1
RVE53256.1 KQ461195 KPJ06661.1 ODYU01006835 SOQ49045.1 AGBW02011497 OWR46556.1 NWSH01000845 PCG73875.1 KZ150017 PZC74957.1 GBGD01002741 JAC86148.1 GBBI01001153 JAC17559.1 PYGN01000257 PSN49920.1 MF942760 AWK28283.1 KK852869 KDR14645.1 GECU01029308 JAS78398.1 GEDC01017850 JAS19448.1 GEZM01003873 JAV96507.1 GBHO01039941 JAG03663.1 GU128093 KQ971379 ACY95476.1 EEZ97330.1 GDHC01014293 JAQ04336.1 NEVH01020330 PNF21871.1 AK417835 BAN21050.1 KM099159 AJZ68820.1 GBHO01039943 GDHC01004434 JAG03661.1 JAQ14195.1 GL446149 EFN88443.1 GL888103 EGI67438.1 ADTU01013618 GGMS01001841 MBY71044.1 GGMR01008731 MBY21350.1 LJIG01022743 KRT78947.1 KQ434867 KZC09150.1 ABLF02036175 KQ981490 KYN41288.1 GAHY01000075 JAA77435.1 KQ414821 KOC60450.1 KQ979999 KYN18318.1 QOIP01000001 RLU27142.1 NNAY01003378 OXU19516.1 KQ976736 KYM76107.1 LBMM01006569 KMQ90501.1 GL764974 EFZ17045.1 GBYB01000105 JAG69872.1 KQ982080 KYQ60235.1 KQ977381 KYN03181.1 KK107260 EZA53994.1 ACPB03005154 KQ435709 KOX80018.1 LJIJ01000399 ODM97884.1 GFDL01005606 JAV29439.1 DS231818 EDS41312.1 CH477594 EAT38548.1 CH477365 EAT42533.1 AK401188 BAM17810.1 KPI95470.1 SOQ49047.1 AK402328 BAM18950.1 GEHC01000070 JAV47575.1 GEZM01082209 JAV61403.1 ATLV01013558 KE524867 KFB38105.1 APCN01004769 APCN01004770 PZC74958.1 AAAB01008811 EAA45375.2 ATLV01013557 KFB38103.1 PCG73874.1 EDS41314.1 GAPW01003911 JAC09687.1 GGMR01001853 MBY14472.1 KDR14646.1 KY031115 ATU82866.1 GFDL01005593 JAV29452.1 PSN49911.1 GEBQ01010488 JAT29489.1 GFXV01004909 MBW16714.1 EAA04933.5 GU128092 ACY95475.1 EEZ97329.1 CH477855 EAT35608.1 GEBQ01020891 JAT19086.1 GEDC01004779 JAS32519.1 GFTR01004118 JAW12308.1 BABH01003008 GAMD01001601 JAA99989.1 MF942759 AWK28282.1 GECZ01021342 JAS48427.1 ADTU01013619 ADTU01013620 GEDC01004272 JAS33026.1
Proteomes
UP000005204
UP000053268
UP000283053
UP000053240
UP000007151
UP000218220
+ More
UP000245037 UP000027135 UP000007266 UP000235965 UP000192223 UP000008237 UP000007755 UP000005205 UP000076502 UP000079169 UP000007819 UP000078541 UP000053825 UP000078492 UP000279307 UP000002358 UP000215335 UP000078540 UP000036403 UP000075809 UP000078542 UP000053097 UP000015103 UP000053105 UP000094527 UP000005203 UP000002320 UP000008820 UP000075903 UP000030765 UP000075840 UP000007062 UP000075880 UP000075900 UP000075920 UP000075901
UP000245037 UP000027135 UP000007266 UP000235965 UP000192223 UP000008237 UP000007755 UP000005205 UP000076502 UP000079169 UP000007819 UP000078541 UP000053825 UP000078492 UP000279307 UP000002358 UP000215335 UP000078540 UP000036403 UP000075809 UP000078542 UP000053097 UP000015103 UP000053105 UP000094527 UP000005203 UP000002320 UP000008820 UP000075903 UP000030765 UP000075840 UP000007062 UP000075880 UP000075900 UP000075920 UP000075901
Pfam
PF01607 CBM_14
SUPFAM
SSF57625
SSF57625
ProteinModelPortal
H9JEF3
I4DJ44
A0A3S2NZP3
A0A194QSZ5
A0A2H1W7G7
A0A212EYI4
+ More
A0A2A4JP85 A0A2W1BNU5 A0A069DPZ6 A0A023F984 A0A2P8Z0B8 A0A2S1ZS45 A0A067QYQ5 A0A1B6HUP9 A0A1B6D180 A0A1Y1NL60 A0A0A9WAZ2 D1MAI5 A0A146LAW5 A0A2J7PZW4 R4WJR8 A0A0F6PN07 A0A1W4XCC5 A0A0A9W8D0 E2B782 F4WEF1 A0A158NER8 A0A2S2Q042 A0A2S2NVZ4 A0A0T6AUY5 A0A154PD68 A0A1S3DGK4 J9JZY1 A0A195FLJ7 R4G5Z9 A0A0L7QP93 A0A195DZL8 A0A3L8E3L9 K7IMV2 A0A232EME2 A0A195AVM1 A0A0J7KJS7 E9IQV8 A0A0C9QWL8 A0A151XIU6 A0A195CR50 A0A026WG30 T1I1T4 A0A0N0BK39 A0A1D2MYL9 A0A087ZT03 A0A1Q3FPB3 B0W0L0 A0A1S4FMS0 Q16VF5 Q178G5 I4DIS0 A0A194PWG1 A0A2H1W7M0 A0A182V3S0 I4DM10 A0A1W7R8Z4 A0A1Y1KJ01 A0A084VJG1 J9K501 A0A1Y9GLC3 A0A2W1BJB4 Q7PFC5 A0A084VJF9 A0A2A4JQJ5 B0W0L2 A0A023EKL3 A0A2S2NBZ9 A0A1S4GAI5 A0A1Y9GLX8 A0A067R898 A0A2K8JM15 A0A1Q3FPL7 A0A2P8Z085 A0A1B6M0M6 A0A2H8TR84 Q7QHZ7 D1MAI4 Q16MN2 A0A1B6L5Z3 A0A1B6E3P5 A0A182J939 A0A224XR67 H9JEF2 T1DJ78 A0A1I8JUT6 A0A182W553 A0A2S1ZS47 A0A1I8JUR0 A0A1B6FE13 A0A182W552 A0A158NER9 A0A1B6E516 A0A182SH98
A0A2A4JP85 A0A2W1BNU5 A0A069DPZ6 A0A023F984 A0A2P8Z0B8 A0A2S1ZS45 A0A067QYQ5 A0A1B6HUP9 A0A1B6D180 A0A1Y1NL60 A0A0A9WAZ2 D1MAI5 A0A146LAW5 A0A2J7PZW4 R4WJR8 A0A0F6PN07 A0A1W4XCC5 A0A0A9W8D0 E2B782 F4WEF1 A0A158NER8 A0A2S2Q042 A0A2S2NVZ4 A0A0T6AUY5 A0A154PD68 A0A1S3DGK4 J9JZY1 A0A195FLJ7 R4G5Z9 A0A0L7QP93 A0A195DZL8 A0A3L8E3L9 K7IMV2 A0A232EME2 A0A195AVM1 A0A0J7KJS7 E9IQV8 A0A0C9QWL8 A0A151XIU6 A0A195CR50 A0A026WG30 T1I1T4 A0A0N0BK39 A0A1D2MYL9 A0A087ZT03 A0A1Q3FPB3 B0W0L0 A0A1S4FMS0 Q16VF5 Q178G5 I4DIS0 A0A194PWG1 A0A2H1W7M0 A0A182V3S0 I4DM10 A0A1W7R8Z4 A0A1Y1KJ01 A0A084VJG1 J9K501 A0A1Y9GLC3 A0A2W1BJB4 Q7PFC5 A0A084VJF9 A0A2A4JQJ5 B0W0L2 A0A023EKL3 A0A2S2NBZ9 A0A1S4GAI5 A0A1Y9GLX8 A0A067R898 A0A2K8JM15 A0A1Q3FPL7 A0A2P8Z085 A0A1B6M0M6 A0A2H8TR84 Q7QHZ7 D1MAI4 Q16MN2 A0A1B6L5Z3 A0A1B6E3P5 A0A182J939 A0A224XR67 H9JEF2 T1DJ78 A0A1I8JUT6 A0A182W553 A0A2S1ZS47 A0A1I8JUR0 A0A1B6FE13 A0A182W552 A0A158NER9 A0A1B6E516 A0A182SH98
PDB
6G9E
E-value=3.48492e-09,
Score=144
Ontologies
GO
Topology
SignalP
Position: 1 - 17,
Likelihood: 0.999717
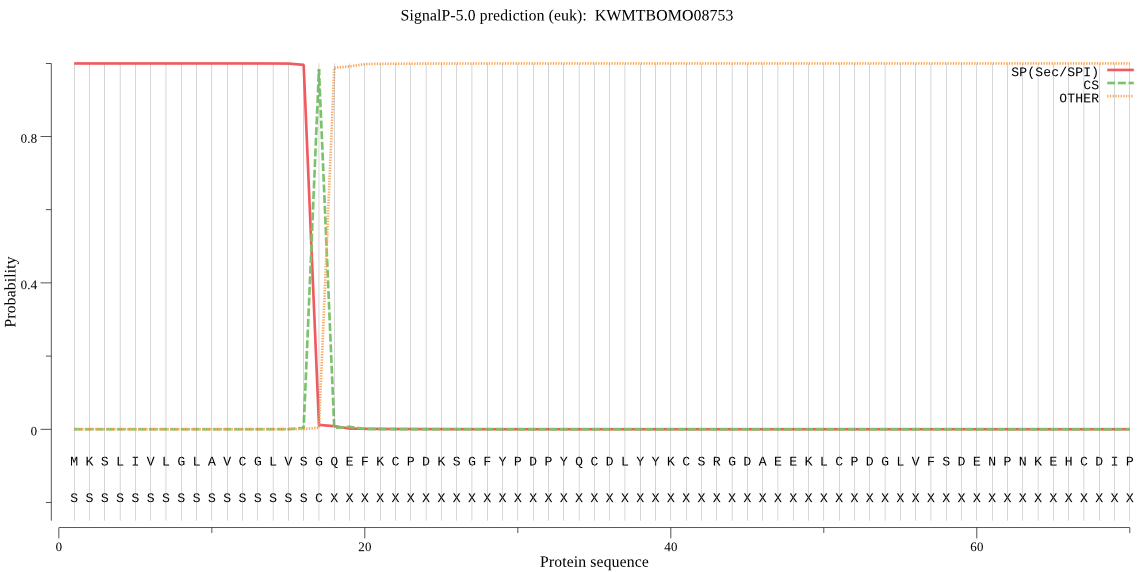
Length:
239
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02067
Exp number, first 60 AAs:
0.02067
Total prob of N-in:
0.01488
outside
1 - 239

Population Genetic Test Statistics
Pi
280.805491
Theta
184.238059
Tajima's D
1.716813
CLR
0.208577
CSRT
0.831458427078646
Interpretation
Uncertain
