Gene
KWMTBOMO08752 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007901
Annotation
PREDICTED:_peritrophin-1-like_[Amyelois_transitella]
Location in the cell
Extracellular Reliability : 4.113
Sequence
CDS
ATGAGTGCAAAGTTCCTTTATCTCGCCGCTTGCGTTGCTTTCGCTAACGCGGGAATACTTCTGGAGCACGCTCCACCATGCCCGGAAGAATACGGTGTCCAGGCATATGCGCATCCCGAGCAGTGCGACCAATTCTTTCTCTGCACTAACGGCACGCTCACCGTAGAGACCTGCGAGAACGGGCTTCTCTTCGACGGAAAGGGTGCAGTTCACAACCACTGCAACTATAATTGGGCTGTTGATTGTGGTCACAGAACAGCGAATTTGGAACCTCTATCAACACCCGGATGCGAATACCAATTCGGTATTTACCCAGATAGCCACGAGTGCTCCACTAGCTACATTAAATGCGCATATGGTGTCCCGGAACAATTTCCCTGCACCCCAGGCTTGGTCTACGATGAGCGCAGTCACAGCTGCAACTGGCCTGATCTCCTCCAACCCTTCTGTAACCCCGAGGCTGTCGTTGGCTTCAAATGCCCATCAAAAGTACCAGCGAACACTCCATCGGCTAAGTTCTGGCCGTTCCCCCGCTTCCCCGTACCTGGAGACTGTCACAGGCTCATCACCTGCGTGGAGGGTCAGCCCCGTCTTATCACCTGCGAAGAAGGAAAAGTGTTCGATGATCAGAACCTGACCTGTGAAGATCCAGAGATCGTTCCTCATTGCGGTCGCGCGTAA
Protein
MSAKFLYLAACVAFANAGILLEHAPPCPEEYGVQAYAHPEQCDQFFLCTNGTLTVETCENGLLFDGKGAVHNHCNYNWAVDCGHRTANLEPLSTPGCEYQFGIYPDSHECSTSYIKCAYGVPEQFPCTPGLVYDERSHSCNWPDLLQPFCNPEAVVGFKCPSKVPANTPSAKFWPFPRFPVPGDCHRLITCVEGQPRLITCEEGKVFDDQNLTCEDPEIVPHCGRA
Summary
Uniprot
H9JEF4
A0A2W1BPZ7
A0A2A4JQ33
A0A212F5E8
A0A2H1VHV2
I4DMX5
+ More
I4DJ81 A0A437BSD3 A0A0L7KVH0 A0A194QNV9 N6UAQ9 D1MAI6 J3JX01 A0A3Q8HGF4 A0A0F6PN44 A0A1W4XB85 A0A2J7PZZ8 F4WEF2 A0A158NER7 A0A067QWT8 A0A151XIK7 A0A195FM05 A0A195E041 A0A195AVW5 A0A182XNG5 A0A182ULI9 A0A0L0BST6 A0A182K897 B4L7X0 A0A182RM35 A0A2P8Z091 A0A2S1ZS43 A0A1Y1K5J6 A0A182PKY2 A0A182VIG4 A0A182W551 A0A182IDQ5 A0A182LIG3 Q5TWB5 A0A1A9Z5U6 A0A1A9XXU1 A0A1B0APQ0 B3MQI2 A0A084VJG2 A0A1B0G8D2 A0A1A9VBM2 A0A182QGH3 A0A182N0U2 A0A182GD29 A0A0A1XSJ5 B4M9Y5 A0A182YK44 A0A336M6J9 A0A023EK87 A0A0M3QZH4 O17452 B0W0L3 B4NBU0 A0A1L8E3D5 A0A154PBD3 T1PG76 A0A1W4UDZ7 B4R2W6 B4PY12 B3NWG9 A0A232EMC5 Q29GW5 W5JCC4 A0A1L8E3Q5 B4I652 R4FNS6 A0A034WTR1 B4JKA1 A0A1Q3FQH5 A0A3B0JD25 W8C900 Q16VF6 A0A0K8US26 A0A1I8P7S7 A0A2A3EBG9 K7IMV1 E9IAA5 A0A1J1HW22 B4GV27 A0A1S4FN36 A0A182J1P5 A0A182MND3 A0A087ZT04 A0A1B6HVH5 A0A1B0DAP4 A0A161MDA6 A0A3L8E4I5 A0A310S879 A0A026WD46 A0A1D2MXV5 A0A224XVN1 A0A0L7QPP6 E0VTN8 R4WE23 A0A0A9WD57
I4DJ81 A0A437BSD3 A0A0L7KVH0 A0A194QNV9 N6UAQ9 D1MAI6 J3JX01 A0A3Q8HGF4 A0A0F6PN44 A0A1W4XB85 A0A2J7PZZ8 F4WEF2 A0A158NER7 A0A067QWT8 A0A151XIK7 A0A195FM05 A0A195E041 A0A195AVW5 A0A182XNG5 A0A182ULI9 A0A0L0BST6 A0A182K897 B4L7X0 A0A182RM35 A0A2P8Z091 A0A2S1ZS43 A0A1Y1K5J6 A0A182PKY2 A0A182VIG4 A0A182W551 A0A182IDQ5 A0A182LIG3 Q5TWB5 A0A1A9Z5U6 A0A1A9XXU1 A0A1B0APQ0 B3MQI2 A0A084VJG2 A0A1B0G8D2 A0A1A9VBM2 A0A182QGH3 A0A182N0U2 A0A182GD29 A0A0A1XSJ5 B4M9Y5 A0A182YK44 A0A336M6J9 A0A023EK87 A0A0M3QZH4 O17452 B0W0L3 B4NBU0 A0A1L8E3D5 A0A154PBD3 T1PG76 A0A1W4UDZ7 B4R2W6 B4PY12 B3NWG9 A0A232EMC5 Q29GW5 W5JCC4 A0A1L8E3Q5 B4I652 R4FNS6 A0A034WTR1 B4JKA1 A0A1Q3FQH5 A0A3B0JD25 W8C900 Q16VF6 A0A0K8US26 A0A1I8P7S7 A0A2A3EBG9 K7IMV1 E9IAA5 A0A1J1HW22 B4GV27 A0A1S4FN36 A0A182J1P5 A0A182MND3 A0A087ZT04 A0A1B6HVH5 A0A1B0DAP4 A0A161MDA6 A0A3L8E4I5 A0A310S879 A0A026WD46 A0A1D2MXV5 A0A224XVN1 A0A0L7QPP6 E0VTN8 R4WE23 A0A0A9WD57
Pubmed
19121390
28756777
22118469
22651552
26354079
26227816
+ More
23537049 18362917 20144715 19820115 22516182 21719571 21347285 24845553 26108605 17994087 29403074 29712872 28004739 20966253 12364791 14747013 17210077 24438588 26483478 25830018 25244985 24945155 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 17550304 28648823 15632085 20920257 23761445 25348373 24495485 17510324 20075255 21282665 30249741 24508170 27289101 20566863 23691247 25401762
23537049 18362917 20144715 19820115 22516182 21719571 21347285 24845553 26108605 17994087 29403074 29712872 28004739 20966253 12364791 14747013 17210077 24438588 26483478 25830018 25244985 24945155 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 17550304 28648823 15632085 20920257 23761445 25348373 24495485 17510324 20075255 21282665 30249741 24508170 27289101 20566863 23691247 25401762
EMBL
BABH01003010
BABH01003011
KZ150017
PZC74956.1
NWSH01000845
PCG73876.1
+ More
AGBW02010205 OWR48929.1 ODYU01002626 SOQ40366.1 AK402643 BAM19265.1 AK401349 KQ459596 BAM17971.1 KPI95468.1 RSAL01000014 RVE53255.1 JTDY01005411 KOB67034.1 KQ461195 KPJ06660.1 APGK01031868 APGK01031869 APGK01031870 APGK01031871 APGK01031872 APGK01031873 KB740829 ENN78820.1 GU128094 KQ971379 ACY95477.1 EEZ97331.1 BT127769 AEE62731.1 MG601670 AYA49995.1 KM099162 AJZ68823.1 NEVH01020330 PNF21898.1 GL888103 EGI67439.1 ADTU01013618 KK852869 KDR14644.1 KQ982080 KYQ60236.1 KQ981490 KYN41287.1 KQ979999 KYN18317.1 KQ976736 KYM76109.1 JRES01001419 KNC23081.1 CH933814 EDW05545.1 PYGN01000257 PSN49919.1 MF942764 AWK28287.1 GEZM01091887 JAV56722.1 APCN01004769 AAAB01008811 EAL41678.2 JXJN01001545 JXJN01001546 JXJN01001547 CH902621 EDV44608.1 ATLV01013559 KE524867 KFB38106.1 CCAG010000242 AXCN02002174 JXUM01055299 KQ561861 KXJ77301.1 GBXI01016528 GBXI01000028 JAC97763.1 JAD14264.1 CH940655 EDW66044.1 UFQT01000526 SSX24991.1 GAPW01004052 JAC09546.1 CP012528 ALC49379.1 AF030895 AE014298 AY069467 AAB86431.1 AAF50926.1 AAL39612.1 AAN09563.1 DS231818 EDS41315.1 CH964239 EDW82299.1 GFDF01000918 JAV13166.1 KQ434867 KZC09151.1 KA646888 AFP61517.1 CM000366 EDX18443.1 CM000162 EDX02984.1 CH954180 EDV46789.1 NNAY01003378 OXU19515.1 CH379064 EAL31994.1 ADMH02001582 ETN61982.1 GFDF01000727 JAV13357.1 CH480823 EDW56258.1 ACPB03005153 GAHY01001174 JAA76336.1 GAKP01001265 JAC57687.1 CH916370 EDW00004.1 GFDL01005218 JAV29827.1 OUUW01000003 SPP78493.1 GAMC01007066 JAB99489.1 CH477594 EAT38547.1 GDHF01022810 JAI29504.1 KZ288312 PBC28401.1 GL761997 EFZ22502.1 CVRI01000020 CRK90710.1 CH479192 EDW26564.1 AXCM01000244 GECU01029081 JAS78625.1 AJVK01004583 GEMB01000889 JAS02251.1 QOIP01000001 RLU27139.1 KQ773121 OAD52431.1 KK107260 EZA53995.1 LJIJ01000399 ODM97883.1 GFTR01003854 JAW12572.1 KQ414821 KOC60451.1 DS235768 EEB16744.1 AK418029 BAN21244.1 GBHO01039161 JAG04443.1
AGBW02010205 OWR48929.1 ODYU01002626 SOQ40366.1 AK402643 BAM19265.1 AK401349 KQ459596 BAM17971.1 KPI95468.1 RSAL01000014 RVE53255.1 JTDY01005411 KOB67034.1 KQ461195 KPJ06660.1 APGK01031868 APGK01031869 APGK01031870 APGK01031871 APGK01031872 APGK01031873 KB740829 ENN78820.1 GU128094 KQ971379 ACY95477.1 EEZ97331.1 BT127769 AEE62731.1 MG601670 AYA49995.1 KM099162 AJZ68823.1 NEVH01020330 PNF21898.1 GL888103 EGI67439.1 ADTU01013618 KK852869 KDR14644.1 KQ982080 KYQ60236.1 KQ981490 KYN41287.1 KQ979999 KYN18317.1 KQ976736 KYM76109.1 JRES01001419 KNC23081.1 CH933814 EDW05545.1 PYGN01000257 PSN49919.1 MF942764 AWK28287.1 GEZM01091887 JAV56722.1 APCN01004769 AAAB01008811 EAL41678.2 JXJN01001545 JXJN01001546 JXJN01001547 CH902621 EDV44608.1 ATLV01013559 KE524867 KFB38106.1 CCAG010000242 AXCN02002174 JXUM01055299 KQ561861 KXJ77301.1 GBXI01016528 GBXI01000028 JAC97763.1 JAD14264.1 CH940655 EDW66044.1 UFQT01000526 SSX24991.1 GAPW01004052 JAC09546.1 CP012528 ALC49379.1 AF030895 AE014298 AY069467 AAB86431.1 AAF50926.1 AAL39612.1 AAN09563.1 DS231818 EDS41315.1 CH964239 EDW82299.1 GFDF01000918 JAV13166.1 KQ434867 KZC09151.1 KA646888 AFP61517.1 CM000366 EDX18443.1 CM000162 EDX02984.1 CH954180 EDV46789.1 NNAY01003378 OXU19515.1 CH379064 EAL31994.1 ADMH02001582 ETN61982.1 GFDF01000727 JAV13357.1 CH480823 EDW56258.1 ACPB03005153 GAHY01001174 JAA76336.1 GAKP01001265 JAC57687.1 CH916370 EDW00004.1 GFDL01005218 JAV29827.1 OUUW01000003 SPP78493.1 GAMC01007066 JAB99489.1 CH477594 EAT38547.1 GDHF01022810 JAI29504.1 KZ288312 PBC28401.1 GL761997 EFZ22502.1 CVRI01000020 CRK90710.1 CH479192 EDW26564.1 AXCM01000244 GECU01029081 JAS78625.1 AJVK01004583 GEMB01000889 JAS02251.1 QOIP01000001 RLU27139.1 KQ773121 OAD52431.1 KK107260 EZA53995.1 LJIJ01000399 ODM97883.1 GFTR01003854 JAW12572.1 KQ414821 KOC60451.1 DS235768 EEB16744.1 AK418029 BAN21244.1 GBHO01039161 JAG04443.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000283053
UP000037510
+ More
UP000053240 UP000019118 UP000007266 UP000192223 UP000235965 UP000007755 UP000005205 UP000027135 UP000075809 UP000078541 UP000078492 UP000078540 UP000076407 UP000075902 UP000037069 UP000075881 UP000009192 UP000075900 UP000245037 UP000075885 UP000075903 UP000075920 UP000075840 UP000075882 UP000007062 UP000092445 UP000092443 UP000092460 UP000007801 UP000030765 UP000092444 UP000078200 UP000075886 UP000075884 UP000069940 UP000249989 UP000008792 UP000076408 UP000092553 UP000000803 UP000002320 UP000007798 UP000076502 UP000095301 UP000192221 UP000000304 UP000002282 UP000008711 UP000215335 UP000001819 UP000000673 UP000001292 UP000015103 UP000001070 UP000268350 UP000008820 UP000095300 UP000242457 UP000002358 UP000183832 UP000008744 UP000075880 UP000075883 UP000005203 UP000092462 UP000279307 UP000053097 UP000094527 UP000053825 UP000009046
UP000053240 UP000019118 UP000007266 UP000192223 UP000235965 UP000007755 UP000005205 UP000027135 UP000075809 UP000078541 UP000078492 UP000078540 UP000076407 UP000075902 UP000037069 UP000075881 UP000009192 UP000075900 UP000245037 UP000075885 UP000075903 UP000075920 UP000075840 UP000075882 UP000007062 UP000092445 UP000092443 UP000092460 UP000007801 UP000030765 UP000092444 UP000078200 UP000075886 UP000075884 UP000069940 UP000249989 UP000008792 UP000076408 UP000092553 UP000000803 UP000002320 UP000007798 UP000076502 UP000095301 UP000192221 UP000000304 UP000002282 UP000008711 UP000215335 UP000001819 UP000000673 UP000001292 UP000015103 UP000001070 UP000268350 UP000008820 UP000095300 UP000242457 UP000002358 UP000183832 UP000008744 UP000075880 UP000075883 UP000005203 UP000092462 UP000279307 UP000053097 UP000094527 UP000053825 UP000009046
Pfam
PF01607 CBM_14
SUPFAM
SSF57625
SSF57625
ProteinModelPortal
H9JEF4
A0A2W1BPZ7
A0A2A4JQ33
A0A212F5E8
A0A2H1VHV2
I4DMX5
+ More
I4DJ81 A0A437BSD3 A0A0L7KVH0 A0A194QNV9 N6UAQ9 D1MAI6 J3JX01 A0A3Q8HGF4 A0A0F6PN44 A0A1W4XB85 A0A2J7PZZ8 F4WEF2 A0A158NER7 A0A067QWT8 A0A151XIK7 A0A195FM05 A0A195E041 A0A195AVW5 A0A182XNG5 A0A182ULI9 A0A0L0BST6 A0A182K897 B4L7X0 A0A182RM35 A0A2P8Z091 A0A2S1ZS43 A0A1Y1K5J6 A0A182PKY2 A0A182VIG4 A0A182W551 A0A182IDQ5 A0A182LIG3 Q5TWB5 A0A1A9Z5U6 A0A1A9XXU1 A0A1B0APQ0 B3MQI2 A0A084VJG2 A0A1B0G8D2 A0A1A9VBM2 A0A182QGH3 A0A182N0U2 A0A182GD29 A0A0A1XSJ5 B4M9Y5 A0A182YK44 A0A336M6J9 A0A023EK87 A0A0M3QZH4 O17452 B0W0L3 B4NBU0 A0A1L8E3D5 A0A154PBD3 T1PG76 A0A1W4UDZ7 B4R2W6 B4PY12 B3NWG9 A0A232EMC5 Q29GW5 W5JCC4 A0A1L8E3Q5 B4I652 R4FNS6 A0A034WTR1 B4JKA1 A0A1Q3FQH5 A0A3B0JD25 W8C900 Q16VF6 A0A0K8US26 A0A1I8P7S7 A0A2A3EBG9 K7IMV1 E9IAA5 A0A1J1HW22 B4GV27 A0A1S4FN36 A0A182J1P5 A0A182MND3 A0A087ZT04 A0A1B6HVH5 A0A1B0DAP4 A0A161MDA6 A0A3L8E4I5 A0A310S879 A0A026WD46 A0A1D2MXV5 A0A224XVN1 A0A0L7QPP6 E0VTN8 R4WE23 A0A0A9WD57
I4DJ81 A0A437BSD3 A0A0L7KVH0 A0A194QNV9 N6UAQ9 D1MAI6 J3JX01 A0A3Q8HGF4 A0A0F6PN44 A0A1W4XB85 A0A2J7PZZ8 F4WEF2 A0A158NER7 A0A067QWT8 A0A151XIK7 A0A195FM05 A0A195E041 A0A195AVW5 A0A182XNG5 A0A182ULI9 A0A0L0BST6 A0A182K897 B4L7X0 A0A182RM35 A0A2P8Z091 A0A2S1ZS43 A0A1Y1K5J6 A0A182PKY2 A0A182VIG4 A0A182W551 A0A182IDQ5 A0A182LIG3 Q5TWB5 A0A1A9Z5U6 A0A1A9XXU1 A0A1B0APQ0 B3MQI2 A0A084VJG2 A0A1B0G8D2 A0A1A9VBM2 A0A182QGH3 A0A182N0U2 A0A182GD29 A0A0A1XSJ5 B4M9Y5 A0A182YK44 A0A336M6J9 A0A023EK87 A0A0M3QZH4 O17452 B0W0L3 B4NBU0 A0A1L8E3D5 A0A154PBD3 T1PG76 A0A1W4UDZ7 B4R2W6 B4PY12 B3NWG9 A0A232EMC5 Q29GW5 W5JCC4 A0A1L8E3Q5 B4I652 R4FNS6 A0A034WTR1 B4JKA1 A0A1Q3FQH5 A0A3B0JD25 W8C900 Q16VF6 A0A0K8US26 A0A1I8P7S7 A0A2A3EBG9 K7IMV1 E9IAA5 A0A1J1HW22 B4GV27 A0A1S4FN36 A0A182J1P5 A0A182MND3 A0A087ZT04 A0A1B6HVH5 A0A1B0DAP4 A0A161MDA6 A0A3L8E4I5 A0A310S879 A0A026WD46 A0A1D2MXV5 A0A224XVN1 A0A0L7QPP6 E0VTN8 R4WE23 A0A0A9WD57
PDB
6G9E
E-value=0.00609392,
Score=90
Ontologies
GO
Topology
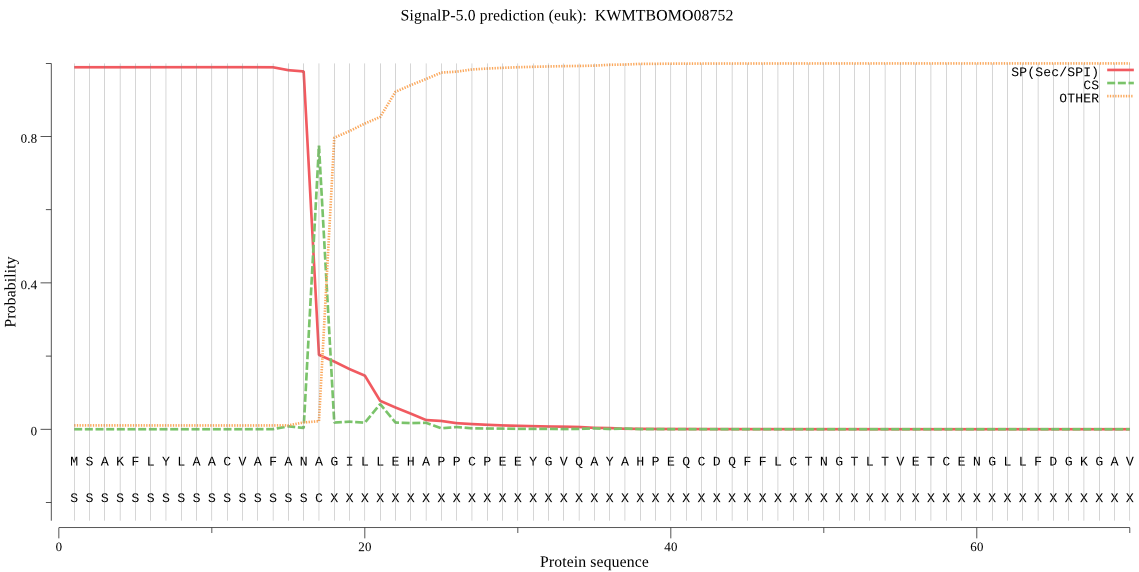
SignalP
Position: 1 - 17,
Likelihood: 0.989430
Length:
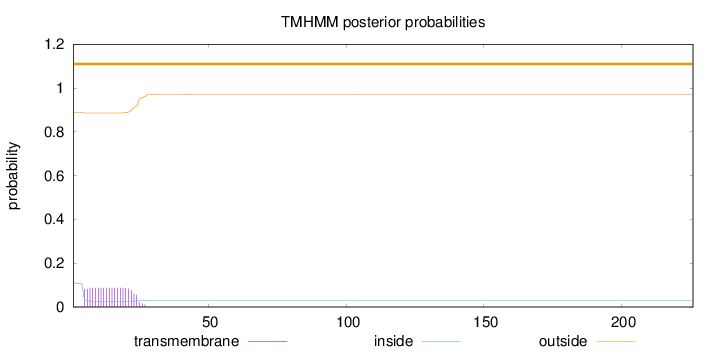
226
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.72681
Exp number, first 60 AAs:
1.72673
Total prob of N-in:
0.11063
outside
1 - 226
Population Genetic Test Statistics
Pi
311.030958
Theta
178.883253
Tajima's D
2.360021
CLR
0.217021
CSRT
0.930203489825509
Interpretation
Uncertain
