Gene
KWMTBOMO08751 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007678
Annotation
probable_chitinase_3_precursor_[Papilio_xuthus]
Location in the cell
Extracellular Reliability : 3.186
Sequence
CDS
ATGGTTAATTTTCGGGTAGCTCTCGCATTTGTAGCGTGCTTTATTGTAGTGACGTCAGCACAGTCACCTAAAGCCAGAAAGAAGCCTGCCAACCCCCCAGCTCGTGCCAGAGAAGAGAGCGAGGCTGACCCGGAGATCACGAACTTATGTCCTGAAGATGGATACTTTGCTGACGCCGAACAGTGTGACAAATATTATGAATGCAGGGGTGGTGATATAATTGAGAAACTTTGCCCAGACGGTATGGTGTTCAACGACTACAGTCCACAGGAAGAGAAATGTGATTTACCTTTCAACCTGGACTGCTCTCAGAGACCCAAGCTTCAAACACCTCAACCTTCCCTCCACTGTATTCGTCAAAATGGCTACTTCTCTCACGAGGATCCTAAGGAATGCGGTAAATTCTACTTTTGCGTTGATGGGAAATTCAACATGATAACCTGTCCTGATGGCTTGGTCTACAACGACAAGACCGGCATCTGCACTTGGCCTGATGAAGCTAAAAAGAAGGGATGTGGCGCTGCAGAGGTATTCCAGTTCGAATGTCCGGCTGTTAATGAAACCTTCGGTCTGACCCACCCTCGGTATGCGGACCCTGACGATTGTCAGTTCTTCTACGTTTGTATCAACGGCAACACCCCCAGGAGATCTGGATGCAAGCTCGGACAGGCCTTTGATGACGTCAGTAAGAAGTGCGAATGGGCTAGGAAAGTTCCTGAATGCGCTGAGTGGTACAAAGGGCAGTTGACTGATGCGGAGTTGGATGCCCTCGAGAACCCACCTACTCCGAAACCCAAGCCTGCTGGTAGTCCTTCGCGCCGGAAGCCGCAACGCCCAAGCAAAGGGAAAACAGTTCAAATAGAAGATGAATAG
Protein
MVNFRVALAFVACFIVVTSAQSPKARKKPANPPARAREESEADPEITNLCPEDGYFADAEQCDKYYECRGGDIIEKLCPDGMVFNDYSPQEEKCDLPFNLDCSQRPKLQTPQPSLHCIRQNGYFSHEDPKECGKFYFCVDGKFNMITCPDGLVYNDKTGICTWPDEAKKKGCGAAEVFQFECPAVNETFGLTHPRYADPDDCQFFYVCINGNTPRRSGCKLGQAFDDVSKKCEWARKVPECAEWYKGQLTDAELDALENPPTPKPKPAGSPSRRKPQRPSKGKTVQIEDE
Summary
Similarity
Belongs to the amiloride-sensitive sodium channel (TC 1.A.6) family.
Uniprot
H9JDT1
A0A194QMT5
I4DJ43
I4DMX3
A0A2H1VHR6
A0A2A4JPA0
+ More
A0A437BS34 A0A212F5C1 A0A2W1BMV3 G9D568 A0A1L8E3R6 Q7Q1H2 A1YR27 A0A182HRT8 A0A182REY1 A0A182WGM5 A0A182T4A6 A0A182KTX9 A0A182GKX7 A0A182VBS9 A0A182P4C8 A0A182NBU9 A0A182QBK0 A0A182YDE8 A0A182F2X9 A0A182KCA1 J3JVT5 T1DSM3 A0A3L8E326 A0A232EMH2 A0A0T6B2G5 A0A1Y1LJC3 A0A158NES0 A0A0F6PMG3 A0A1S4FYV3 Q16JT9 A0A195FKU7 A0A182J0I3 E2B784 A0A2H8TMJ0 A0A0M9AAQ6 F4WEE9 A0A154PFN7 W5JFH9 B0WGT7 A0A310SJI6 A0A2S2QCF2 A0A2A3E870 J9JUR7 A0A067QWD6 A0A0J7L0S5 A0A2J7PZW1 A0A1B6BWG6 A0A2S1ZS48 A0A0K8TBE4 A0A0A9Y9N7 E9IAA7 A0A1I8P8P7 T1PAF5 A0A1B6D8C4 A0A1I8MWD8 B3MPD4 A0A026WG30 A0A3Q8HG55 A0A0A1X4V5 B4KH47 E0VTN7 A0A034WUX0 B4N3E8 A0A087ZR24 A0A0L0CNF2 A0A0K8W266 A0A195AVM1 A0A3B0JU29 A0A1B6KYR4 A0A1B6INL7 B4M9E3 Q29NG5 B4G7M8 A0A146M7S2 W8C4V7 Q9Y156 B4HWF2 B4Q8C4 A0A151XIU6 A0A0M3QTM4 B3N915 B4NYR8 A0A1W4UFH1 B4JCH8 A0A195CR50 A0A1A9XN49 A0A1B0AT80 A0A1J1IV86 A0A1B0AJQ9 A0A1B0FLZ7 A0A1A9UPL5 A0A084WG74
A0A437BS34 A0A212F5C1 A0A2W1BMV3 G9D568 A0A1L8E3R6 Q7Q1H2 A1YR27 A0A182HRT8 A0A182REY1 A0A182WGM5 A0A182T4A6 A0A182KTX9 A0A182GKX7 A0A182VBS9 A0A182P4C8 A0A182NBU9 A0A182QBK0 A0A182YDE8 A0A182F2X9 A0A182KCA1 J3JVT5 T1DSM3 A0A3L8E326 A0A232EMH2 A0A0T6B2G5 A0A1Y1LJC3 A0A158NES0 A0A0F6PMG3 A0A1S4FYV3 Q16JT9 A0A195FKU7 A0A182J0I3 E2B784 A0A2H8TMJ0 A0A0M9AAQ6 F4WEE9 A0A154PFN7 W5JFH9 B0WGT7 A0A310SJI6 A0A2S2QCF2 A0A2A3E870 J9JUR7 A0A067QWD6 A0A0J7L0S5 A0A2J7PZW1 A0A1B6BWG6 A0A2S1ZS48 A0A0K8TBE4 A0A0A9Y9N7 E9IAA7 A0A1I8P8P7 T1PAF5 A0A1B6D8C4 A0A1I8MWD8 B3MPD4 A0A026WG30 A0A3Q8HG55 A0A0A1X4V5 B4KH47 E0VTN7 A0A034WUX0 B4N3E8 A0A087ZR24 A0A0L0CNF2 A0A0K8W266 A0A195AVM1 A0A3B0JU29 A0A1B6KYR4 A0A1B6INL7 B4M9E3 Q29NG5 B4G7M8 A0A146M7S2 W8C4V7 Q9Y156 B4HWF2 B4Q8C4 A0A151XIU6 A0A0M3QTM4 B3N915 B4NYR8 A0A1W4UFH1 B4JCH8 A0A195CR50 A0A1A9XN49 A0A1B0AT80 A0A1J1IV86 A0A1B0AJQ9 A0A1B0FLZ7 A0A1A9UPL5 A0A084WG74
Pubmed
19121390
26354079
22651552
22118469
28756777
21967427
+ More
12364791 14747013 17210077 18362917 19820115 20966253 26483478 25244985 22516182 23537049 30249741 28648823 28004739 21347285 17510324 20798317 21719571 20920257 23761445 24845553 29712872 26823975 25401762 21282665 25315136 17994087 18057021 24508170 25830018 20566863 25348373 26108605 15632085 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 17550304 24438588
12364791 14747013 17210077 18362917 19820115 20966253 26483478 25244985 22516182 23537049 30249741 28648823 28004739 21347285 17510324 20798317 21719571 20920257 23761445 24845553 29712872 26823975 25401762 21282665 25315136 17994087 18057021 24508170 25830018 20566863 25348373 26108605 15632085 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 17550304 24438588
EMBL
BABH01003011
BABH01003012
KQ461195
KPJ06659.1
AK401311
KQ459596
+ More
BAM17933.1 KPI95467.1 AK402641 BAM19263.1 ODYU01002626 SOQ40368.1 NWSH01000845 PCG73877.1 RSAL01000014 RVE53254.1 AGBW02010205 OWR48930.1 KZ150017 PZC74955.1 JF777471 AEU11816.1 GFDF01000704 JAV13380.1 AAAB01008980 EAA14163.4 EF125544 KQ971379 ABL73928.1 EEZ97328.1 APCN01001747 JXUM01013416 JXUM01013417 KQ560376 KXJ82723.1 AXCN02000691 APGK01055130 APGK01055131 BT127353 KB741259 KB631924 AEE62315.1 ENN71734.1 ERL87186.1 GAMD01001414 JAB00177.1 QOIP01000001 RLU27140.1 NNAY01003378 OXU19518.1 LJIG01016112 KRT81578.1 GEZM01057072 JAV72460.1 ADTU01013620 KM099160 AJZ68821.1 CH477993 EAT34543.1 KQ981490 KYN41290.1 GL446149 EFN88445.1 GFXV01003582 MBW15387.1 KQ435709 KOX79966.1 GL888103 EGI67436.1 KQ434893 KZC10669.1 ADMH02001647 ETN61564.1 DS231929 EDS27192.1 KQ759804 OAD62821.1 GGMS01006190 MBY75393.1 KZ288344 PBC27704.1 ABLF02036175 KK852869 KDR14648.1 LBMM01001484 KMQ96231.1 NEVH01020330 PNF21873.1 GEDC01031666 JAS05632.1 MF942761 AWK28284.1 GBRD01003090 GDHC01001609 JAG62731.1 JAQ17020.1 GBHO01017369 JAG26235.1 GL761997 EFZ22495.1 KA645756 AFP60385.1 GEDC01015398 JAS21900.1 CH902620 EDV32253.2 KPU73860.1 KK107260 EZA53994.1 MG601671 AYA49996.1 GBXI01008574 JAD05718.1 CH933807 EDW12258.1 DS235768 EEB16743.1 GAKP01000830 JAC58122.1 CH964085 EDW78887.2 JRES01000143 KNC33850.1 GDHF01025646 GDHF01010296 GDHF01007125 JAI26668.1 JAI42018.1 JAI45189.1 KQ976736 KYM76107.1 OUUW01000010 SPP85617.1 GEBQ01023389 GEBQ01004789 JAT16588.1 JAT35188.1 GECU01019184 JAS88522.1 CH940654 EDW57819.1 CH379060 EAL33377.3 CH479180 EDW28428.1 GDHC01003823 JAQ14806.1 GAMC01009346 JAB97209.1 AF145612 AE014134 AAD38587.1 AAF52851.1 CH480818 EDW52347.1 CM000361 CM002910 EDX04446.1 KMY89392.1 KQ982080 KYQ60235.1 CP012523 ALC39139.1 CH954177 EDV58450.1 CM000157 EDW88732.1 CH916368 EDW03132.1 KQ977381 KYN03181.1 JXJN01003202 JXJN01003203 CVRI01000061 CRL04125.1 CCAG010007147 ATLV01023460 KE525343 KFB49218.1
BAM17933.1 KPI95467.1 AK402641 BAM19263.1 ODYU01002626 SOQ40368.1 NWSH01000845 PCG73877.1 RSAL01000014 RVE53254.1 AGBW02010205 OWR48930.1 KZ150017 PZC74955.1 JF777471 AEU11816.1 GFDF01000704 JAV13380.1 AAAB01008980 EAA14163.4 EF125544 KQ971379 ABL73928.1 EEZ97328.1 APCN01001747 JXUM01013416 JXUM01013417 KQ560376 KXJ82723.1 AXCN02000691 APGK01055130 APGK01055131 BT127353 KB741259 KB631924 AEE62315.1 ENN71734.1 ERL87186.1 GAMD01001414 JAB00177.1 QOIP01000001 RLU27140.1 NNAY01003378 OXU19518.1 LJIG01016112 KRT81578.1 GEZM01057072 JAV72460.1 ADTU01013620 KM099160 AJZ68821.1 CH477993 EAT34543.1 KQ981490 KYN41290.1 GL446149 EFN88445.1 GFXV01003582 MBW15387.1 KQ435709 KOX79966.1 GL888103 EGI67436.1 KQ434893 KZC10669.1 ADMH02001647 ETN61564.1 DS231929 EDS27192.1 KQ759804 OAD62821.1 GGMS01006190 MBY75393.1 KZ288344 PBC27704.1 ABLF02036175 KK852869 KDR14648.1 LBMM01001484 KMQ96231.1 NEVH01020330 PNF21873.1 GEDC01031666 JAS05632.1 MF942761 AWK28284.1 GBRD01003090 GDHC01001609 JAG62731.1 JAQ17020.1 GBHO01017369 JAG26235.1 GL761997 EFZ22495.1 KA645756 AFP60385.1 GEDC01015398 JAS21900.1 CH902620 EDV32253.2 KPU73860.1 KK107260 EZA53994.1 MG601671 AYA49996.1 GBXI01008574 JAD05718.1 CH933807 EDW12258.1 DS235768 EEB16743.1 GAKP01000830 JAC58122.1 CH964085 EDW78887.2 JRES01000143 KNC33850.1 GDHF01025646 GDHF01010296 GDHF01007125 JAI26668.1 JAI42018.1 JAI45189.1 KQ976736 KYM76107.1 OUUW01000010 SPP85617.1 GEBQ01023389 GEBQ01004789 JAT16588.1 JAT35188.1 GECU01019184 JAS88522.1 CH940654 EDW57819.1 CH379060 EAL33377.3 CH479180 EDW28428.1 GDHC01003823 JAQ14806.1 GAMC01009346 JAB97209.1 AF145612 AE014134 AAD38587.1 AAF52851.1 CH480818 EDW52347.1 CM000361 CM002910 EDX04446.1 KMY89392.1 KQ982080 KYQ60235.1 CP012523 ALC39139.1 CH954177 EDV58450.1 CM000157 EDW88732.1 CH916368 EDW03132.1 KQ977381 KYN03181.1 JXJN01003202 JXJN01003203 CVRI01000061 CRL04125.1 CCAG010007147 ATLV01023460 KE525343 KFB49218.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000283053
UP000007151
+ More
UP000007062 UP000007266 UP000075840 UP000075900 UP000075920 UP000075901 UP000075882 UP000069940 UP000249989 UP000075903 UP000075885 UP000075884 UP000075886 UP000076408 UP000069272 UP000075881 UP000019118 UP000030742 UP000279307 UP000215335 UP000005205 UP000008820 UP000078541 UP000075880 UP000008237 UP000053105 UP000007755 UP000076502 UP000000673 UP000002320 UP000242457 UP000007819 UP000027135 UP000036403 UP000235965 UP000095300 UP000095301 UP000007801 UP000053097 UP000009192 UP000009046 UP000007798 UP000005203 UP000037069 UP000078540 UP000268350 UP000008792 UP000001819 UP000008744 UP000000803 UP000001292 UP000000304 UP000075809 UP000092553 UP000008711 UP000002282 UP000192221 UP000001070 UP000078542 UP000092443 UP000092460 UP000183832 UP000092445 UP000092444 UP000078200 UP000030765
UP000007062 UP000007266 UP000075840 UP000075900 UP000075920 UP000075901 UP000075882 UP000069940 UP000249989 UP000075903 UP000075885 UP000075884 UP000075886 UP000076408 UP000069272 UP000075881 UP000019118 UP000030742 UP000279307 UP000215335 UP000005205 UP000008820 UP000078541 UP000075880 UP000008237 UP000053105 UP000007755 UP000076502 UP000000673 UP000002320 UP000242457 UP000007819 UP000027135 UP000036403 UP000235965 UP000095300 UP000095301 UP000007801 UP000053097 UP000009192 UP000009046 UP000007798 UP000005203 UP000037069 UP000078540 UP000268350 UP000008792 UP000001819 UP000008744 UP000000803 UP000001292 UP000000304 UP000075809 UP000092553 UP000008711 UP000002282 UP000192221 UP000001070 UP000078542 UP000092443 UP000092460 UP000183832 UP000092445 UP000092444 UP000078200 UP000030765
SUPFAM
SSF57625
SSF57625
Gene 3D
ProteinModelPortal
H9JDT1
A0A194QMT5
I4DJ43
I4DMX3
A0A2H1VHR6
A0A2A4JPA0
+ More
A0A437BS34 A0A212F5C1 A0A2W1BMV3 G9D568 A0A1L8E3R6 Q7Q1H2 A1YR27 A0A182HRT8 A0A182REY1 A0A182WGM5 A0A182T4A6 A0A182KTX9 A0A182GKX7 A0A182VBS9 A0A182P4C8 A0A182NBU9 A0A182QBK0 A0A182YDE8 A0A182F2X9 A0A182KCA1 J3JVT5 T1DSM3 A0A3L8E326 A0A232EMH2 A0A0T6B2G5 A0A1Y1LJC3 A0A158NES0 A0A0F6PMG3 A0A1S4FYV3 Q16JT9 A0A195FKU7 A0A182J0I3 E2B784 A0A2H8TMJ0 A0A0M9AAQ6 F4WEE9 A0A154PFN7 W5JFH9 B0WGT7 A0A310SJI6 A0A2S2QCF2 A0A2A3E870 J9JUR7 A0A067QWD6 A0A0J7L0S5 A0A2J7PZW1 A0A1B6BWG6 A0A2S1ZS48 A0A0K8TBE4 A0A0A9Y9N7 E9IAA7 A0A1I8P8P7 T1PAF5 A0A1B6D8C4 A0A1I8MWD8 B3MPD4 A0A026WG30 A0A3Q8HG55 A0A0A1X4V5 B4KH47 E0VTN7 A0A034WUX0 B4N3E8 A0A087ZR24 A0A0L0CNF2 A0A0K8W266 A0A195AVM1 A0A3B0JU29 A0A1B6KYR4 A0A1B6INL7 B4M9E3 Q29NG5 B4G7M8 A0A146M7S2 W8C4V7 Q9Y156 B4HWF2 B4Q8C4 A0A151XIU6 A0A0M3QTM4 B3N915 B4NYR8 A0A1W4UFH1 B4JCH8 A0A195CR50 A0A1A9XN49 A0A1B0AT80 A0A1J1IV86 A0A1B0AJQ9 A0A1B0FLZ7 A0A1A9UPL5 A0A084WG74
A0A437BS34 A0A212F5C1 A0A2W1BMV3 G9D568 A0A1L8E3R6 Q7Q1H2 A1YR27 A0A182HRT8 A0A182REY1 A0A182WGM5 A0A182T4A6 A0A182KTX9 A0A182GKX7 A0A182VBS9 A0A182P4C8 A0A182NBU9 A0A182QBK0 A0A182YDE8 A0A182F2X9 A0A182KCA1 J3JVT5 T1DSM3 A0A3L8E326 A0A232EMH2 A0A0T6B2G5 A0A1Y1LJC3 A0A158NES0 A0A0F6PMG3 A0A1S4FYV3 Q16JT9 A0A195FKU7 A0A182J0I3 E2B784 A0A2H8TMJ0 A0A0M9AAQ6 F4WEE9 A0A154PFN7 W5JFH9 B0WGT7 A0A310SJI6 A0A2S2QCF2 A0A2A3E870 J9JUR7 A0A067QWD6 A0A0J7L0S5 A0A2J7PZW1 A0A1B6BWG6 A0A2S1ZS48 A0A0K8TBE4 A0A0A9Y9N7 E9IAA7 A0A1I8P8P7 T1PAF5 A0A1B6D8C4 A0A1I8MWD8 B3MPD4 A0A026WG30 A0A3Q8HG55 A0A0A1X4V5 B4KH47 E0VTN7 A0A034WUX0 B4N3E8 A0A087ZR24 A0A0L0CNF2 A0A0K8W266 A0A195AVM1 A0A3B0JU29 A0A1B6KYR4 A0A1B6INL7 B4M9E3 Q29NG5 B4G7M8 A0A146M7S2 W8C4V7 Q9Y156 B4HWF2 B4Q8C4 A0A151XIU6 A0A0M3QTM4 B3N915 B4NYR8 A0A1W4UFH1 B4JCH8 A0A195CR50 A0A1A9XN49 A0A1B0AT80 A0A1J1IV86 A0A1B0AJQ9 A0A1B0FLZ7 A0A1A9UPL5 A0A084WG74
PDB
6G9E
E-value=2.66164e-09,
Score=147
Ontologies
GO
PANTHER
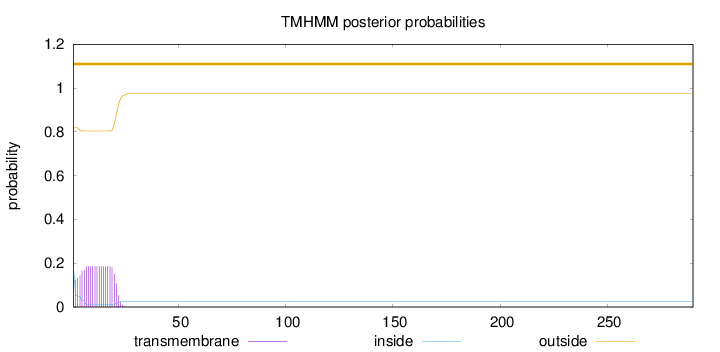
Topology
SignalP
Position: 1 - 20,
Likelihood: 0.990568

Length:
290
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.49764
Exp number, first 60 AAs:
3.49764
Total prob of N-in:
0.18134
outside
1 - 290
Population Genetic Test Statistics
Pi
275.747778
Theta
28.504974
Tajima's D
0.313672
CLR
1.699297
CSRT
0.464876756162192
Interpretation
Uncertain
