Gene
KWMTBOMO08747 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007677
Annotation
gasp_precursor_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 3.693
Sequence
CDS
ATGAATTCGAGGTGTTTATTAGTGTTAGCGGCCGCCCTCGGCGTAGCTGTGGCCCAAGAGTCCTTCAAATGCCCCGACGACTTCGGTTTCTACCCTCACCACATCTCCTGTGACAAATACTGGAAGTGCGACAACGGTGTAGCCGAACTGAAAACCTGCGGCAACGGTCTCGCTTTCGACGCCACAGATTCCAAATACTTGACCGAGAACTGCGACTACCTTCACAACGTTGAGTGCGGAGAGAGGACACAGCTTGAGCCCCCAATCTCCACTCCTCACTGTTCGAGACTGTACGGCATCTTCCCTGATGAAAACAAGTGCGACGTTTTCTGGAACTGCTGGAACGGAGAAGCCTCCCGCTATCAGTGCAGCCCCGGACTTGCCTACGACAGAGAGTCCCGCGTCTGCATGTGGGCTGATCAGGTTCCGGAGTGCAAAAACGAAGAAGTAGCAAACGGATTCGGCTGCCCAGCCCCCGGTGAGGTCTCCAACGCTGGCTCGTTCAGCCGTCACGCTCATCCCGAAGATTGCCGTAAATATTACATCTGTCTCGAGGGCGTTGCCCGCGAGTACGGTTGCCCCATCGGAACCGTTTTCAAGATCGGAGACGCCGACGGCACTGGTAACTGCGAAGACCCCGAAGACGTTCCCGGATGCGAGGACTACTATGGAGAATTGGATCTGAAGGCGATCCGCAAGAGTGAGCTCCTCGCCGGGCTCCAGGCTGACGGCCAGCCTCGCCCCAACCAGCCCAAGCAGCTCAAACCCCGTCCTCCAAAGGAGAACTAA
Protein
MNSRCLLVLAAALGVAVAQESFKCPDDFGFYPHHISCDKYWKCDNGVAELKTCGNGLAFDATDSKYLTENCDYLHNVECGERTQLEPPISTPHCSRLYGIFPDENKCDVFWNCWNGEASRYQCSPGLAYDRESRVCMWADQVPECKNEEVANGFGCPAPGEVSNAGSFSRHAHPEDCRKYYICLEGVAREYGCPIGTVFKIGDADGTGNCEDPEDVPGCEDYYGELDLKAIRKSELLAGLQADGQPRPNQPKQLKPRPPKEN
Summary
Uniprot
H9JDT0
Q1HPN5
A0A2A4K7D0
A0A2H1VHQ4
I4DLV5
I4DIC0
+ More
A0A194PWF6 A0A212F5B6 A0A0L7KWR9 G9D533 A0A182NL79 A0A182QH04 A0A182HJG9 A0A182YQC4 A0A2W1BIR3 A0A2A4K6W0 A1YR28 A0A182V0X4 A0A182LCU0 A0A182PKS0 F5HM59 A0A182VQK4 C5MRD6 A0A182RQS4 A0A139WN65 B0WJP1 A0A182XKH5 A0A1Q3FR70 Q297X8 B4G369 B4MBS1 A0A3L8E417 A0A3B0K9K5 B4JYH0 A0A1I8P396 Q9VNL0 B4QXN5 E2B778 B3NYK5 Q16I79 A0A0M4F4S4 T1P893 W5JI62 A0A182FAV7 A0A1W4UR95 A0A182JIN4 A0A182KE30 B4NHF0 B3LWA7 B4I4A8 B4PVL1 A0A0L0C0Q9 B4KB02 A0A158NER5 A0A194QMA9 A0A0J7L4D4 A0A0A1WHY2 A0A195FLJ1 A0A0M9ABR1 A0A034VQ79 A0A195AVM7 G9D540 A0A3S2M7I5 A0A1J1HK29 A0A336MR94 W8BI37 A0A0K8W7B0 D3TRR5 A0A1B0A1L2 A0A0N8ER11 A0A087ZR01 E0VGD4 A0A310S521 A0A222AJ11 A0A154PB79 A0A232EJ67 A0A3B0JCW8 A0A0R3NH60 Q7PKA3 A0A2A3EB14 A0A0Q9WBZ2 A0A023EKK0 A8JQU3 A0A1D1XPE6 A0A0Q9X4U2 T1PMI6 A0A0P9A158 A0A0R1E8X8 A0A1B6J5D0 A0A0L7QP90 A0A026WDF3 A0A0Q9X0U0 A0A1B0CRW7 A0A195CR45 A0A1B0G939 A0A1L8E3Q9 A0A0K8V4B4 A1YR29 A0A2J7QFJ7 A0A1S3CWB5
A0A194PWF6 A0A212F5B6 A0A0L7KWR9 G9D533 A0A182NL79 A0A182QH04 A0A182HJG9 A0A182YQC4 A0A2W1BIR3 A0A2A4K6W0 A1YR28 A0A182V0X4 A0A182LCU0 A0A182PKS0 F5HM59 A0A182VQK4 C5MRD6 A0A182RQS4 A0A139WN65 B0WJP1 A0A182XKH5 A0A1Q3FR70 Q297X8 B4G369 B4MBS1 A0A3L8E417 A0A3B0K9K5 B4JYH0 A0A1I8P396 Q9VNL0 B4QXN5 E2B778 B3NYK5 Q16I79 A0A0M4F4S4 T1P893 W5JI62 A0A182FAV7 A0A1W4UR95 A0A182JIN4 A0A182KE30 B4NHF0 B3LWA7 B4I4A8 B4PVL1 A0A0L0C0Q9 B4KB02 A0A158NER5 A0A194QMA9 A0A0J7L4D4 A0A0A1WHY2 A0A195FLJ1 A0A0M9ABR1 A0A034VQ79 A0A195AVM7 G9D540 A0A3S2M7I5 A0A1J1HK29 A0A336MR94 W8BI37 A0A0K8W7B0 D3TRR5 A0A1B0A1L2 A0A0N8ER11 A0A087ZR01 E0VGD4 A0A310S521 A0A222AJ11 A0A154PB79 A0A232EJ67 A0A3B0JCW8 A0A0R3NH60 Q7PKA3 A0A2A3EB14 A0A0Q9WBZ2 A0A023EKK0 A8JQU3 A0A1D1XPE6 A0A0Q9X4U2 T1PMI6 A0A0P9A158 A0A0R1E8X8 A0A1B6J5D0 A0A0L7QP90 A0A026WDF3 A0A0Q9X0U0 A0A1B0CRW7 A0A195CR45 A0A1B0G939 A0A1L8E3Q9 A0A0K8V4B4 A1YR29 A0A2J7QFJ7 A0A1S3CWB5
Pubmed
19121390
22651552
26354079
22118469
26227816
21967427
+ More
25244985 28756777 18362917 19820115 20966253 12364791 14747013 17210077 20043914 15632085 17994087 30249741 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20798317 17510324 25315136 20920257 23761445 17550304 26108605 21347285 25830018 25348373 24495485 20353571 20566863 28648823 24945155 24508170
25244985 28756777 18362917 19820115 20966253 12364791 14747013 17210077 20043914 15632085 17994087 30249741 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20798317 17510324 25315136 20920257 23761445 17550304 26108605 21347285 25830018 25348373 24495485 20353571 20566863 28648823 24945155 24508170
EMBL
BABH01003014
DQ443367
ABF51456.1
NWSH01000070
PCG79979.1
ODYU01002626
+ More
SOQ40370.1 AK402273 BAM18895.1 AK401038 BAM17660.1 KQ459596 KPI95465.1 AGBW02010205 OWR48932.1 JTDY01004839 KOB67688.1 JF777436 AEU11781.1 AXCN02000074 AXCN02000075 APCN01002164 KZ150017 PZC74952.1 PCG79977.1 EF125545 KQ971312 ABL73929.1 KYB29334.1 AAAB01008982 EGK97370.1 GQ144964 ACR61397.1 KYB29333.1 DS231962 EDS29368.1 GFDL01005020 JAV30025.1 CM000070 EAL28077.2 CH479179 EDW24264.1 CH940656 EDW58542.1 QOIP01000001 RLU27366.1 OUUW01000005 SPP80228.1 CH916377 EDV90732.1 AE014297 AY069336 AAF51920.1 AAL39481.1 CM000364 EDX11821.1 GL446149 EFN88439.1 CH954181 EDV47986.1 CH478100 EAT33969.1 CP012526 ALC46420.1 KA644814 AFP59443.1 ADMH02001333 ETN62973.1 CH964272 EDW84626.1 CH902617 EDV43740.1 CH480821 EDW55051.1 CM000160 EDW97820.1 JRES01001160 KNC25044.1 CH933806 EDW14680.1 ADTU01013612 ADTU01013613 ADTU01013614 KQ461195 KPJ06657.1 LBMM01000831 KMQ97403.1 GBXI01015815 JAC98476.1 KQ981490 KYN41283.1 KQ435709 KOX80013.1 GAKP01014705 JAC44247.1 KQ976736 KYM76112.1 JF777443 AEU11788.1 RSAL01000014 RVE53252.1 CVRI01000006 CRK88275.1 UFQT01001957 SSX32225.1 GAMC01008143 JAB98412.1 GDHF01005375 JAI46939.1 EZ424117 ADD20393.1 GDIQ01002380 JAN92357.1 DS235138 EEB12440.1 KQ773121 OAD52427.1 KY563862 ASO76353.1 KQ434867 KZC09155.1 NNAY01004076 OXU18406.1 SPP80227.1 KRT00425.1 EAA43448.5 KZ288312 PBC28399.1 KRF78334.1 GAPW01003895 JAC09703.1 ABW08608.1 GDJX01023666 JAT44270.1 KRG00082.1 KA649143 AFP63772.1 KPU80438.1 KRK03988.1 GECU01013328 JAS94378.1 KQ414821 KOC60453.1 KK107260 EZA53998.1 KRG01145.1 AJWK01025386 KQ977381 KYN03176.1 CCAG010009318 GFDF01000872 JAV13212.1 GDHF01018572 JAI33742.1 EF125546 ABL73930.1 KYB29332.1 NEVH01015297 PNF27352.1
SOQ40370.1 AK402273 BAM18895.1 AK401038 BAM17660.1 KQ459596 KPI95465.1 AGBW02010205 OWR48932.1 JTDY01004839 KOB67688.1 JF777436 AEU11781.1 AXCN02000074 AXCN02000075 APCN01002164 KZ150017 PZC74952.1 PCG79977.1 EF125545 KQ971312 ABL73929.1 KYB29334.1 AAAB01008982 EGK97370.1 GQ144964 ACR61397.1 KYB29333.1 DS231962 EDS29368.1 GFDL01005020 JAV30025.1 CM000070 EAL28077.2 CH479179 EDW24264.1 CH940656 EDW58542.1 QOIP01000001 RLU27366.1 OUUW01000005 SPP80228.1 CH916377 EDV90732.1 AE014297 AY069336 AAF51920.1 AAL39481.1 CM000364 EDX11821.1 GL446149 EFN88439.1 CH954181 EDV47986.1 CH478100 EAT33969.1 CP012526 ALC46420.1 KA644814 AFP59443.1 ADMH02001333 ETN62973.1 CH964272 EDW84626.1 CH902617 EDV43740.1 CH480821 EDW55051.1 CM000160 EDW97820.1 JRES01001160 KNC25044.1 CH933806 EDW14680.1 ADTU01013612 ADTU01013613 ADTU01013614 KQ461195 KPJ06657.1 LBMM01000831 KMQ97403.1 GBXI01015815 JAC98476.1 KQ981490 KYN41283.1 KQ435709 KOX80013.1 GAKP01014705 JAC44247.1 KQ976736 KYM76112.1 JF777443 AEU11788.1 RSAL01000014 RVE53252.1 CVRI01000006 CRK88275.1 UFQT01001957 SSX32225.1 GAMC01008143 JAB98412.1 GDHF01005375 JAI46939.1 EZ424117 ADD20393.1 GDIQ01002380 JAN92357.1 DS235138 EEB12440.1 KQ773121 OAD52427.1 KY563862 ASO76353.1 KQ434867 KZC09155.1 NNAY01004076 OXU18406.1 SPP80227.1 KRT00425.1 EAA43448.5 KZ288312 PBC28399.1 KRF78334.1 GAPW01003895 JAC09703.1 ABW08608.1 GDJX01023666 JAT44270.1 KRG00082.1 KA649143 AFP63772.1 KPU80438.1 KRK03988.1 GECU01013328 JAS94378.1 KQ414821 KOC60453.1 KK107260 EZA53998.1 KRG01145.1 AJWK01025386 KQ977381 KYN03176.1 CCAG010009318 GFDF01000872 JAV13212.1 GDHF01018572 JAI33742.1 EF125546 ABL73930.1 KYB29332.1 NEVH01015297 PNF27352.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000037510
UP000075884
+ More
UP000075886 UP000075840 UP000076408 UP000007266 UP000075903 UP000075882 UP000075885 UP000007062 UP000075920 UP000075900 UP000002320 UP000076407 UP000001819 UP000008744 UP000008792 UP000279307 UP000268350 UP000001070 UP000095300 UP000000803 UP000000304 UP000008237 UP000008711 UP000008820 UP000092553 UP000095301 UP000000673 UP000069272 UP000192221 UP000075880 UP000075881 UP000007798 UP000007801 UP000001292 UP000002282 UP000037069 UP000009192 UP000005205 UP000053240 UP000036403 UP000078541 UP000053105 UP000078540 UP000283053 UP000183832 UP000092445 UP000005203 UP000009046 UP000076502 UP000215335 UP000242457 UP000053825 UP000053097 UP000092461 UP000078542 UP000092444 UP000235965 UP000079169
UP000075886 UP000075840 UP000076408 UP000007266 UP000075903 UP000075882 UP000075885 UP000007062 UP000075920 UP000075900 UP000002320 UP000076407 UP000001819 UP000008744 UP000008792 UP000279307 UP000268350 UP000001070 UP000095300 UP000000803 UP000000304 UP000008237 UP000008711 UP000008820 UP000092553 UP000095301 UP000000673 UP000069272 UP000192221 UP000075880 UP000075881 UP000007798 UP000007801 UP000001292 UP000002282 UP000037069 UP000009192 UP000005205 UP000053240 UP000036403 UP000078541 UP000053105 UP000078540 UP000283053 UP000183832 UP000092445 UP000005203 UP000009046 UP000076502 UP000215335 UP000242457 UP000053825 UP000053097 UP000092461 UP000078542 UP000092444 UP000235965 UP000079169
Pfam
PF01607 CBM_14
SUPFAM
SSF57625
SSF57625
ProteinModelPortal
H9JDT0
Q1HPN5
A0A2A4K7D0
A0A2H1VHQ4
I4DLV5
I4DIC0
+ More
A0A194PWF6 A0A212F5B6 A0A0L7KWR9 G9D533 A0A182NL79 A0A182QH04 A0A182HJG9 A0A182YQC4 A0A2W1BIR3 A0A2A4K6W0 A1YR28 A0A182V0X4 A0A182LCU0 A0A182PKS0 F5HM59 A0A182VQK4 C5MRD6 A0A182RQS4 A0A139WN65 B0WJP1 A0A182XKH5 A0A1Q3FR70 Q297X8 B4G369 B4MBS1 A0A3L8E417 A0A3B0K9K5 B4JYH0 A0A1I8P396 Q9VNL0 B4QXN5 E2B778 B3NYK5 Q16I79 A0A0M4F4S4 T1P893 W5JI62 A0A182FAV7 A0A1W4UR95 A0A182JIN4 A0A182KE30 B4NHF0 B3LWA7 B4I4A8 B4PVL1 A0A0L0C0Q9 B4KB02 A0A158NER5 A0A194QMA9 A0A0J7L4D4 A0A0A1WHY2 A0A195FLJ1 A0A0M9ABR1 A0A034VQ79 A0A195AVM7 G9D540 A0A3S2M7I5 A0A1J1HK29 A0A336MR94 W8BI37 A0A0K8W7B0 D3TRR5 A0A1B0A1L2 A0A0N8ER11 A0A087ZR01 E0VGD4 A0A310S521 A0A222AJ11 A0A154PB79 A0A232EJ67 A0A3B0JCW8 A0A0R3NH60 Q7PKA3 A0A2A3EB14 A0A0Q9WBZ2 A0A023EKK0 A8JQU3 A0A1D1XPE6 A0A0Q9X4U2 T1PMI6 A0A0P9A158 A0A0R1E8X8 A0A1B6J5D0 A0A0L7QP90 A0A026WDF3 A0A0Q9X0U0 A0A1B0CRW7 A0A195CR45 A0A1B0G939 A0A1L8E3Q9 A0A0K8V4B4 A1YR29 A0A2J7QFJ7 A0A1S3CWB5
A0A194PWF6 A0A212F5B6 A0A0L7KWR9 G9D533 A0A182NL79 A0A182QH04 A0A182HJG9 A0A182YQC4 A0A2W1BIR3 A0A2A4K6W0 A1YR28 A0A182V0X4 A0A182LCU0 A0A182PKS0 F5HM59 A0A182VQK4 C5MRD6 A0A182RQS4 A0A139WN65 B0WJP1 A0A182XKH5 A0A1Q3FR70 Q297X8 B4G369 B4MBS1 A0A3L8E417 A0A3B0K9K5 B4JYH0 A0A1I8P396 Q9VNL0 B4QXN5 E2B778 B3NYK5 Q16I79 A0A0M4F4S4 T1P893 W5JI62 A0A182FAV7 A0A1W4UR95 A0A182JIN4 A0A182KE30 B4NHF0 B3LWA7 B4I4A8 B4PVL1 A0A0L0C0Q9 B4KB02 A0A158NER5 A0A194QMA9 A0A0J7L4D4 A0A0A1WHY2 A0A195FLJ1 A0A0M9ABR1 A0A034VQ79 A0A195AVM7 G9D540 A0A3S2M7I5 A0A1J1HK29 A0A336MR94 W8BI37 A0A0K8W7B0 D3TRR5 A0A1B0A1L2 A0A0N8ER11 A0A087ZR01 E0VGD4 A0A310S521 A0A222AJ11 A0A154PB79 A0A232EJ67 A0A3B0JCW8 A0A0R3NH60 Q7PKA3 A0A2A3EB14 A0A0Q9WBZ2 A0A023EKK0 A8JQU3 A0A1D1XPE6 A0A0Q9X4U2 T1PMI6 A0A0P9A158 A0A0R1E8X8 A0A1B6J5D0 A0A0L7QP90 A0A026WDF3 A0A0Q9X0U0 A0A1B0CRW7 A0A195CR45 A0A1B0G939 A0A1L8E3Q9 A0A0K8V4B4 A1YR29 A0A2J7QFJ7 A0A1S3CWB5
PDB
6G9E
E-value=1.61115e-05,
Score=113
Ontologies
GO
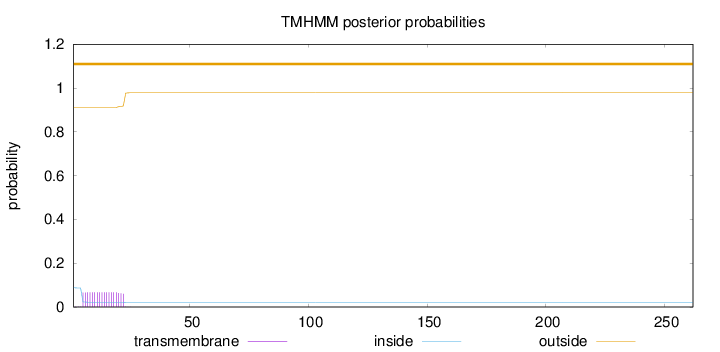
Topology
SignalP
Position: 1 - 18,
Likelihood: 0.999691

Length:
262
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.20192
Exp number, first 60 AAs:
1.20192
Total prob of N-in:
0.08860
outside
1 - 262
Population Genetic Test Statistics
Pi
187.522111
Theta
174.25483
Tajima's D
0.130053
CLR
1.704563
CSRT
0.407529623518824
Interpretation
Uncertain
