Gene
KWMTBOMO08744
Pre Gene Modal
BGIBMGA007903
Annotation
PREDICTED:_tigger_transposable_element-derived_protein_6-like_[Acyrthosiphon_pisum]
Location in the cell
Nuclear Reliability : 2.67
Sequence
CDS
ATGGCGTTCAGGACTAAACGCAAAGCATTTTCATTTCACGATAAATTGAAAATATTACAATATTACGACAAAAAATCACAAGATTACAAAAAATCTCAAATAGCAGCTGAACTTGAAATTCCTGTCTCAACACTATCGACGATTGTAAAATACAGGGCTGATGTAGAAAATAGATGTAAAAACATAAGTGGTGCATACTTAAAACGAAAGAAAAATAAAAGCGGTAAATATGTTGAAATTGAGAATGTTTTGATCGACTGGATCGTCAGTATGAACGCCCAGAATATTCCCCTGGACGGGCCAATGATGAAACAAAGAGCTTTAGAAATCGCTAAAGAATTAGAAGTCTACGATTTCACGGCTTCCAGTGGTTGGTACAGTCGTTTTAAGCAGCGCCACGCGTTCTCGTTAAACAATGACTTTGACAGTGATGACGGCTCCGACCTTACTTACGAATCCTATCAAGAACTAGTCAAAGACTATGAACCGTGTGATGTTTTTAATGCTGATGAATTCGGATTGTTTTACAAGCTAATGCCAAACGACATGTCTACTATAGTAGACAAGATATGTAAAGATGGTACAATGCCAGAAGAACGGTTCACAGTATTGGCATGTTCGAATTCTATTGGATCAGAAAAATTACCACTACTCATTATTGGTAATACTGAAAATTCAGATAAAGACTTGCCCTGCCAATATCAAGTTAACCCTAAGTCTTGGATGACAAAACTGGCTTTCACAAACTGGTTATACATTCTTGACGAATATTTTGAAATTCAGGGGAGATTGGTGGTATTGTTCGTCGATGAGAGTCCAGTGCATCCAAAGGACATAATCTTGAATAACATAAACTTGGTTATCATACCCCCTGAAGCTGAACATGAAATGCCTATGTCTCAAGGCATTATTAAGGTTATTAAGCAGAATTATAGGATCAAATTGATTGAAAAATACAAGGAGGGACTAGCTGTTGATTTTAAACAGTGCTTGAAATACCTCACTGATGTCTTTGAGTCCATTAAAGCTCATACAATTCAATCTTATTTTAAGAAATCCGGTTTCGGAGACTATTATGAGGTTGATGACGAGGATGACGAATACAAAGAACAAATACTGATGAAATACCAAGACTATTTGTATGTTGATTATGAATTAGCGACGTGTGAAGTAGAAAAAGGCGTTAACCAAGAATCAACATTTACAGATAATAATATAAAAGAAAAAGTTAATAAAATCATCATTAACAATGGTGAAACCATTGATAACAAATCAGTTACGGCGCTTCAAGCGGTCACTACATTGAAGCATTATTTAAAATCTTTAGATTCTGAAAATGGTCTCAACAATTTGTCTGTAATAGAAAGTATAATTTTAAGCTCGATTTCTAAAAATAATTCTATGTAG
Protein
MAFRTKRKAFSFHDKLKILQYYDKKSQDYKKSQIAAELEIPVSTLSTIVKYRADVENRCKNISGAYLKRKKNKSGKYVEIENVLIDWIVSMNAQNIPLDGPMMKQRALEIAKELEVYDFTASSGWYSRFKQRHAFSLNNDFDSDDGSDLTYESYQELVKDYEPCDVFNADEFGLFYKLMPNDMSTIVDKICKDGTMPEERFTVLACSNSIGSEKLPLLIIGNTENSDKDLPCQYQVNPKSWMTKLAFTNWLYILDEYFEIQGRLVVLFVDESPVHPKDIILNNINLVIIPPEAEHEMPMSQGIIKVIKQNYRIKLIEKYKEGLAVDFKQCLKYLTDVFESIKAHTIQSYFKKSGFGDYYEVDDEDDEYKEQILMKYQDYLYVDYELATCEVEKGVNQESTFTDNNIKEKVNKIIINNGETIDNKSVTALQAVTTLKHYLKSLDSENGLNNLSVIESIILSSISKNNSM
Summary
Uniprot
H9JEF6
J9LIK9
J9JIZ7
A0A0V1LHS6
A0A0V1C573
A0A1Y3EAG5
+ More
A0A2S2Q986 A0A0V1CHI8 A0A2H4EWE8 L7MDS5 A0A023GKV9 A0A147BBA7 L7MCT9 A0A087TT27 L7MME9 L7MKK8 A0A1E1XRR1 A0A2H4EU06 A0A1Q3EU22 A0A131XNC6 A0A1E1X2Z9 A0A2R5L9X9 A0A131XM02 A0A1W4WFJ1 L7MM78 L7MAM5 A0A131Z8V0 A0A1U7RJI3 A0A151PB98 G3MP65 A0A1E1XNV7 A0A1E1XIT7 A0A1E1XTJ5 A0A1E1X418 A0A093HII8 A0A224Z1D2 A0A2S2QVQ0 J9K6L9 A0A1E1X318 F6WAY4 J9K454 A0A2S2PW91 L7MLD5 A0A147BJV1 F6WUM2 A0A224Z9W5 A0A3B3SSQ9 A0A147BDF0 L7MMS1 L7MCM4 A0A1E1X3N4 A0A2R5LGG9 A0A1S3A562 J9KW05 A0A1E1X420 A0A2S2PVR1 A0A2R5L8Q6 I3MYQ0 A0A2S2PEI6 A0A1E1X2Z8 F6SYA9 A0A023GKX0 A0A1B6LVP4 G3WWJ9 D2H8M5 A0A1E1X313 L7MLS7 A0A452FPK3 A0A3Q7NNY9 A0A384C8E4 A0A2Y9HTM8 A0A2U3YD76 A0A2U3W4A0 A0A1U7TKK1 H0WRQ3 A0A3Q7WN60 A0A2K6JVL4 A0A2K6NL06 A0A2K5H7G9 A0A452SIC2 A0A2Y9DPY4 K7EXN0 L7MKX2 M3Z6I0 A0A2Y9FY04 A0A2S2QI15 G3W0T3 A0A1E1X307 A0A2K5L488 W5Q0B7
A0A2S2Q986 A0A0V1CHI8 A0A2H4EWE8 L7MDS5 A0A023GKV9 A0A147BBA7 L7MCT9 A0A087TT27 L7MME9 L7MKK8 A0A1E1XRR1 A0A2H4EU06 A0A1Q3EU22 A0A131XNC6 A0A1E1X2Z9 A0A2R5L9X9 A0A131XM02 A0A1W4WFJ1 L7MM78 L7MAM5 A0A131Z8V0 A0A1U7RJI3 A0A151PB98 G3MP65 A0A1E1XNV7 A0A1E1XIT7 A0A1E1XTJ5 A0A1E1X418 A0A093HII8 A0A224Z1D2 A0A2S2QVQ0 J9K6L9 A0A1E1X318 F6WAY4 J9K454 A0A2S2PW91 L7MLD5 A0A147BJV1 F6WUM2 A0A224Z9W5 A0A3B3SSQ9 A0A147BDF0 L7MMS1 L7MCM4 A0A1E1X3N4 A0A2R5LGG9 A0A1S3A562 J9KW05 A0A1E1X420 A0A2S2PVR1 A0A2R5L8Q6 I3MYQ0 A0A2S2PEI6 A0A1E1X2Z8 F6SYA9 A0A023GKX0 A0A1B6LVP4 G3WWJ9 D2H8M5 A0A1E1X313 L7MLS7 A0A452FPK3 A0A3Q7NNY9 A0A384C8E4 A0A2Y9HTM8 A0A2U3YD76 A0A2U3W4A0 A0A1U7TKK1 H0WRQ3 A0A3Q7WN60 A0A2K6JVL4 A0A2K6NL06 A0A2K5H7G9 A0A452SIC2 A0A2Y9DPY4 K7EXN0 L7MKX2 M3Z6I0 A0A2Y9FY04 A0A2S2QI15 G3W0T3 A0A1E1X307 A0A2K5L488 W5Q0B7
Pubmed
EMBL
BABH01003018
ABLF02042388
ABLF02007575
JYDW01000048
KRZ59035.1
JYDI01000635
+ More
KRY44322.1 LVZM01022273 OUC40836.1 GGMS01005095 MBY74298.1 JYDI01000198 KRY48745.1 KU056809 AOE48153.1 GACK01003641 JAA61393.1 GBBM01000897 JAC34521.1 GEGO01007356 JAR88048.1 GACK01004010 JAA61024.1 KK116622 KFM68266.1 GACK01000525 JAA64509.1 GACK01000524 JAA64510.1 GFAA01001803 JAU01632.1 KU056810 AOE48154.1 GFDL01016236 JAV18809.1 GEFH01000749 JAP67832.1 GFAC01005543 JAT93645.1 GGLE01002176 MBY06302.1 GEFH01000408 JAP68173.1 GACK01000576 JAA64458.1 GACK01003708 JAA61326.1 GEDV01001661 JAP86896.1 AKHW03000533 KYO46313.1 JO843666 AEO35283.1 GFAA01002466 JAU00969.1 GFAC01000040 JAT99148.1 GFAA01000830 JAU02605.1 GFAC01005196 JAT93992.1 KL206458 KFV82448.1 GFPF01012512 MAA23658.1 GGMS01012636 MBY81839.1 ABLF02022231 GFAC01005545 JAT93643.1 ABLF02032381 ABLF02032385 GGMS01000470 MBY69673.1 GACK01000229 JAA64805.1 GEGO01004400 JAR91004.1 AAMC01120866 GFPF01012384 MAA23530.1 GEGO01006601 JAR88803.1 GACK01000230 JAA64804.1 GACK01004075 JAA60959.1 GFAC01005542 JAT93646.1 GGLE01004497 MBY08623.1 ABLF02020128 ABLF02020129 GFAC01005195 JAT93993.1 GGMS01000424 MBY69627.1 GGLE01001733 MBY05859.1 AGTP01094548 GGMR01015193 MBY27812.1 GFAC01005549 JAT93639.1 GBBM01000959 JAC34459.1 GEBQ01012216 JAT27761.1 AEFK01016728 ACTA01192650 GL192580 EFB21249.1 GFAC01005556 JAT93632.1 GACK01000074 JAA64960.1 LWLT01000018 AAQR03075137 AGCU01009440 GACK01000434 JAA64600.1 AEYP01000962 GGMS01008154 MBY77357.1 AEFK01215205 GFAC01005555 JAT93633.1 AMGL01037157
KRY44322.1 LVZM01022273 OUC40836.1 GGMS01005095 MBY74298.1 JYDI01000198 KRY48745.1 KU056809 AOE48153.1 GACK01003641 JAA61393.1 GBBM01000897 JAC34521.1 GEGO01007356 JAR88048.1 GACK01004010 JAA61024.1 KK116622 KFM68266.1 GACK01000525 JAA64509.1 GACK01000524 JAA64510.1 GFAA01001803 JAU01632.1 KU056810 AOE48154.1 GFDL01016236 JAV18809.1 GEFH01000749 JAP67832.1 GFAC01005543 JAT93645.1 GGLE01002176 MBY06302.1 GEFH01000408 JAP68173.1 GACK01000576 JAA64458.1 GACK01003708 JAA61326.1 GEDV01001661 JAP86896.1 AKHW03000533 KYO46313.1 JO843666 AEO35283.1 GFAA01002466 JAU00969.1 GFAC01000040 JAT99148.1 GFAA01000830 JAU02605.1 GFAC01005196 JAT93992.1 KL206458 KFV82448.1 GFPF01012512 MAA23658.1 GGMS01012636 MBY81839.1 ABLF02022231 GFAC01005545 JAT93643.1 ABLF02032381 ABLF02032385 GGMS01000470 MBY69673.1 GACK01000229 JAA64805.1 GEGO01004400 JAR91004.1 AAMC01120866 GFPF01012384 MAA23530.1 GEGO01006601 JAR88803.1 GACK01000230 JAA64804.1 GACK01004075 JAA60959.1 GFAC01005542 JAT93646.1 GGLE01004497 MBY08623.1 ABLF02020128 ABLF02020129 GFAC01005195 JAT93993.1 GGMS01000424 MBY69627.1 GGLE01001733 MBY05859.1 AGTP01094548 GGMR01015193 MBY27812.1 GFAC01005549 JAT93639.1 GBBM01000959 JAC34459.1 GEBQ01012216 JAT27761.1 AEFK01016728 ACTA01192650 GL192580 EFB21249.1 GFAC01005556 JAT93632.1 GACK01000074 JAA64960.1 LWLT01000018 AAQR03075137 AGCU01009440 GACK01000434 JAA64600.1 AEYP01000962 GGMS01008154 MBY77357.1 AEFK01215205 GFAC01005555 JAT93633.1 AMGL01037157
Proteomes
UP000005204
UP000007819
UP000054721
UP000054653
UP000243006
UP000054359
+ More
UP000192223 UP000189705 UP000050525 UP000053584 UP000002279 UP000008143 UP000261540 UP000079721 UP000005215 UP000002280 UP000007648 UP000008912 UP000291000 UP000286641 UP000261680 UP000291021 UP000248481 UP000245341 UP000245340 UP000189704 UP000005225 UP000286642 UP000233180 UP000233200 UP000233080 UP000291022 UP000248480 UP000007267 UP000000715 UP000248484 UP000233060 UP000002356
UP000192223 UP000189705 UP000050525 UP000053584 UP000002279 UP000008143 UP000261540 UP000079721 UP000005215 UP000002280 UP000007648 UP000008912 UP000291000 UP000286641 UP000261680 UP000291021 UP000248481 UP000245341 UP000245340 UP000189704 UP000005225 UP000286642 UP000233180 UP000233200 UP000233080 UP000291022 UP000248480 UP000007267 UP000000715 UP000248484 UP000233060 UP000002356
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JEF6
J9LIK9
J9JIZ7
A0A0V1LHS6
A0A0V1C573
A0A1Y3EAG5
+ More
A0A2S2Q986 A0A0V1CHI8 A0A2H4EWE8 L7MDS5 A0A023GKV9 A0A147BBA7 L7MCT9 A0A087TT27 L7MME9 L7MKK8 A0A1E1XRR1 A0A2H4EU06 A0A1Q3EU22 A0A131XNC6 A0A1E1X2Z9 A0A2R5L9X9 A0A131XM02 A0A1W4WFJ1 L7MM78 L7MAM5 A0A131Z8V0 A0A1U7RJI3 A0A151PB98 G3MP65 A0A1E1XNV7 A0A1E1XIT7 A0A1E1XTJ5 A0A1E1X418 A0A093HII8 A0A224Z1D2 A0A2S2QVQ0 J9K6L9 A0A1E1X318 F6WAY4 J9K454 A0A2S2PW91 L7MLD5 A0A147BJV1 F6WUM2 A0A224Z9W5 A0A3B3SSQ9 A0A147BDF0 L7MMS1 L7MCM4 A0A1E1X3N4 A0A2R5LGG9 A0A1S3A562 J9KW05 A0A1E1X420 A0A2S2PVR1 A0A2R5L8Q6 I3MYQ0 A0A2S2PEI6 A0A1E1X2Z8 F6SYA9 A0A023GKX0 A0A1B6LVP4 G3WWJ9 D2H8M5 A0A1E1X313 L7MLS7 A0A452FPK3 A0A3Q7NNY9 A0A384C8E4 A0A2Y9HTM8 A0A2U3YD76 A0A2U3W4A0 A0A1U7TKK1 H0WRQ3 A0A3Q7WN60 A0A2K6JVL4 A0A2K6NL06 A0A2K5H7G9 A0A452SIC2 A0A2Y9DPY4 K7EXN0 L7MKX2 M3Z6I0 A0A2Y9FY04 A0A2S2QI15 G3W0T3 A0A1E1X307 A0A2K5L488 W5Q0B7
A0A2S2Q986 A0A0V1CHI8 A0A2H4EWE8 L7MDS5 A0A023GKV9 A0A147BBA7 L7MCT9 A0A087TT27 L7MME9 L7MKK8 A0A1E1XRR1 A0A2H4EU06 A0A1Q3EU22 A0A131XNC6 A0A1E1X2Z9 A0A2R5L9X9 A0A131XM02 A0A1W4WFJ1 L7MM78 L7MAM5 A0A131Z8V0 A0A1U7RJI3 A0A151PB98 G3MP65 A0A1E1XNV7 A0A1E1XIT7 A0A1E1XTJ5 A0A1E1X418 A0A093HII8 A0A224Z1D2 A0A2S2QVQ0 J9K6L9 A0A1E1X318 F6WAY4 J9K454 A0A2S2PW91 L7MLD5 A0A147BJV1 F6WUM2 A0A224Z9W5 A0A3B3SSQ9 A0A147BDF0 L7MMS1 L7MCM4 A0A1E1X3N4 A0A2R5LGG9 A0A1S3A562 J9KW05 A0A1E1X420 A0A2S2PVR1 A0A2R5L8Q6 I3MYQ0 A0A2S2PEI6 A0A1E1X2Z8 F6SYA9 A0A023GKX0 A0A1B6LVP4 G3WWJ9 D2H8M5 A0A1E1X313 L7MLS7 A0A452FPK3 A0A3Q7NNY9 A0A384C8E4 A0A2Y9HTM8 A0A2U3YD76 A0A2U3W4A0 A0A1U7TKK1 H0WRQ3 A0A3Q7WN60 A0A2K6JVL4 A0A2K6NL06 A0A2K5H7G9 A0A452SIC2 A0A2Y9DPY4 K7EXN0 L7MKX2 M3Z6I0 A0A2Y9FY04 A0A2S2QI15 G3W0T3 A0A1E1X307 A0A2K5L488 W5Q0B7
PDB
1HLV
E-value=2.33516e-15,
Score=201
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus
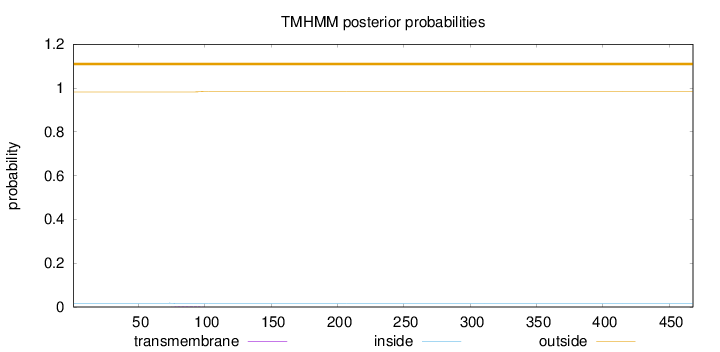
Length:
468
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0390500000000001
Exp number, first 60 AAs:
0.00096
Total prob of N-in:
0.01761
outside
1 - 468
Population Genetic Test Statistics
Pi
149.845727
Theta
152.964625
Tajima's D
0.325579
CLR
13.399977
CSRT
0.463626818659067
Interpretation
Uncertain
