Gene
KWMTBOMO08742
Pre Gene Modal
BGIBMGA007676
Annotation
PREDICTED:_U4/U6.U5_tri-snRNP-associated_protein_2_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 1.95
Sequence
CDS
ATGACTGAAGAACAATCTAGAAAACGCAAAATAACTGATGAAAATGGGACTGACAAACAAGGACGATATTCGAAAACTCAACAAATTCAATGTCCTTACCTGGATACTATCAATCGGCATGTTTTAGATTTTGACTTTGAGAAATTGTGCTCGGTTTCATTGACTAGAATCAATGTTTACGCTTGCTTAGTGTGCGGTAAATATTTTCAGGGAAGAGGCACCAACACTCATGCCTACACACATTCAGTGGCTGATGGACACCATGTATTTTTGAATCTTAACACATTAAAGTTCTATTGTCTTCCTGACAATTATGAAGTTATTGACTCTTCATTGAATGACATCAAATATGTTTTGAACCCAACATTCACACCTGAACAGATCAAGCTTATAGATACTAACACAAAGATGTCACGAGCCATTGATGGCACAATGTATATGCCTGGCATTGTAGGCCTTAATAATATCAAGGCCAATGATTACTGTAATGTAATTTTGCAGTGTTTGTCACAAGTACGTCCCCTACGTAACTACTTTTTGAGAGAAGAGAACTATGCTAGTGTAAAAAGACCACCAGGGGACTCGTCTTTTTTACTTGTACAAAGATTTGGTGAATTATTACGCAAACTATGGAACCCCCGTGCTTTTAAAGCACATGTATCTCCTCACGAGATGTTGCAGGCTGTTGTGCTGTGGTCTAAGAAGCGATATCAGTTTATTAAGCAGAGTGATCCAATAGACTTTCTGTCTTGGTTTCTAAATTCATTACACATGGCACTGAATGGTACAAAGAAGCCAAATAGCTCCATAATTTATAAGTCATTTCTGGGTCACATGAGGATCTATACAAGAAAGTTGCCACCGCCGGATGCTGATGATGCAGCTAAAGTCGATCTTAGCAGTGAAGAGTACAATGAAATGATCACTGAGTCTCCATTTTTGTACCTCACATGTGATTTGCCTCCAACGCCATTGTTCACTGATGAGTTCCGCGAAAATATTATTCCACAGGTAAATCTTTACCAGCTGTTGTCAAAATTCAATGGTCAAACATCAAAAGAATACAAGACTTATAAAGAGAACTTTTTGAAAAGATTTGAAATAACACAGCTGCCTCCTTATCTAATTCTTTACATAAAAAGGTTTACGAAAAATACTTTTTTCGTTGAAAAGAATCCTACAGTTGTAAACTTTCCTGTCAAAAACGTCGATTTTGGCGATATACTGACTGCGGAAATTAAGGCGAAACATAACGGAAAAACAATATACGAACTGGTTGGTAACATTGTTCATGATGGAACACCTGAGAAAGGGACATACAGAGCTCACGTGTTGCATAAGCCTACACAGCAGTGGTATGAGATGCAAGATCTTCATGTTACCAGTATTTTACCTCAAATGATTACACTTACTGAAGCATATATACAAATATATGAACTAAAGCAGGACTAA
Protein
MTEEQSRKRKITDENGTDKQGRYSKTQQIQCPYLDTINRHVLDFDFEKLCSVSLTRINVYACLVCGKYFQGRGTNTHAYTHSVADGHHVFLNLNTLKFYCLPDNYEVIDSSLNDIKYVLNPTFTPEQIKLIDTNTKMSRAIDGTMYMPGIVGLNNIKANDYCNVILQCLSQVRPLRNYFLREENYASVKRPPGDSSFLLVQRFGELLRKLWNPRAFKAHVSPHEMLQAVVLWSKKRYQFIKQSDPIDFLSWFLNSLHMALNGTKKPNSSIIYKSFLGHMRIYTRKLPPPDADDAAKVDLSSEEYNEMITESPFLYLTCDLPPTPLFTDEFRENIIPQVNLYQLLSKFNGQTSKEYKTYKENFLKRFEITQLPPYLILYIKRFTKNTFFVEKNPTVVNFPVKNVDFGDILTAEIKAKHNGKTIYELVGNIVHDGTPEKGTYRAHVLHKPTQQWYEMQDLHVTSILPQMITLTEAYIQIYELKQD
Summary
Uniprot
H9JDS9
A0A2H1VHW3
A0A2A4K864
A0A2W1BIS2
S4P7I5
A0A437BST3
+ More
A0A194PPT9 A0A212F4J7 A0A1E1WA46 D6WET8 A0A151I745 F4WBM0 A0A195BD01 A0A195FSV0 A0A158NRL6 A0A151X5N3 A0A067RII4 A0A1B6ER25 E9J8N0 A0A2J7QQB8 A0A1B6K4Z3 E2A6G5 A0A1Y1LEJ8 E2B4N5 A0A1B6DHZ2 A0A026WQY7 A0A2A3ER45 A0A0L7R947 A0A195DII0 A0A088AFY9 N6UBQ8 E0VSX7 A0A0T6AXA6 A0A154PDK1 A0A232FAM9 A0A0N0U7V5 A0A0A9W861 K7J134 A0A069DU87 A0A232F4U6 A0A0V0G8Y6 A0A2P8YQR4 A0A2S2PZU1 A0A224XPL3 J9JPK8 T1HFN5 W8BZI9 A0A1J1I3J9 A0A1L8E5F3 A0A1B0FDZ7 A0A1A9VRS2 A0A1B0AN93 A0A1A9WSX4 Q17K27 U5ERX4 A0A3B0KE61 A0A182QCP5 A0A182GYZ3 A0A182GIQ4 A0A084WH76 A0A1Q3EWS9 A0A1S4GY37 B3MZV5 A0A182L0K3 A0A182ULV8 A0A182XFY0 A0A182IC66 A0A1W4U9X5 Q7Q4G1 B4PWG5 A0A182TTI1 A0A182PLR3 B3NUP8 A0A182IPX3 A0A182K645 A0A182RDR3 A0A034W1P3 B5DNL3 A0A2M4AT25 A0A182NTZ6 A0A0R3P1D0 A0A182YDU5 A0A0K8TZ59 B0W4M2 A0A182F3F9 A0A182MHL0 A0A1B0GMV9 A0A182W6F9 B4L5I7 A0A0A1WYF5 B4M2Z9 W5JS46 A0A0M4FAC4 A0A1I8P9J7 A0A1I8P9J3 A0A1I8MYQ0 A0A1I8MYP6 B4MTF3 B4I6Y4 Q9VWP1
A0A194PPT9 A0A212F4J7 A0A1E1WA46 D6WET8 A0A151I745 F4WBM0 A0A195BD01 A0A195FSV0 A0A158NRL6 A0A151X5N3 A0A067RII4 A0A1B6ER25 E9J8N0 A0A2J7QQB8 A0A1B6K4Z3 E2A6G5 A0A1Y1LEJ8 E2B4N5 A0A1B6DHZ2 A0A026WQY7 A0A2A3ER45 A0A0L7R947 A0A195DII0 A0A088AFY9 N6UBQ8 E0VSX7 A0A0T6AXA6 A0A154PDK1 A0A232FAM9 A0A0N0U7V5 A0A0A9W861 K7J134 A0A069DU87 A0A232F4U6 A0A0V0G8Y6 A0A2P8YQR4 A0A2S2PZU1 A0A224XPL3 J9JPK8 T1HFN5 W8BZI9 A0A1J1I3J9 A0A1L8E5F3 A0A1B0FDZ7 A0A1A9VRS2 A0A1B0AN93 A0A1A9WSX4 Q17K27 U5ERX4 A0A3B0KE61 A0A182QCP5 A0A182GYZ3 A0A182GIQ4 A0A084WH76 A0A1Q3EWS9 A0A1S4GY37 B3MZV5 A0A182L0K3 A0A182ULV8 A0A182XFY0 A0A182IC66 A0A1W4U9X5 Q7Q4G1 B4PWG5 A0A182TTI1 A0A182PLR3 B3NUP8 A0A182IPX3 A0A182K645 A0A182RDR3 A0A034W1P3 B5DNL3 A0A2M4AT25 A0A182NTZ6 A0A0R3P1D0 A0A182YDU5 A0A0K8TZ59 B0W4M2 A0A182F3F9 A0A182MHL0 A0A1B0GMV9 A0A182W6F9 B4L5I7 A0A0A1WYF5 B4M2Z9 W5JS46 A0A0M4FAC4 A0A1I8P9J7 A0A1I8P9J3 A0A1I8MYQ0 A0A1I8MYP6 B4MTF3 B4I6Y4 Q9VWP1
Pubmed
19121390
28756777
23622113
26354079
22118469
18362917
+ More
19820115 21719571 21347285 24845553 21282665 20798317 28004739 24508170 30249741 23537049 20566863 28648823 25401762 26823975 20075255 26334808 29403074 24495485 17510324 26483478 24438588 12364791 17994087 20966253 17550304 25348373 15632085 25244985 25830018 20920257 23761445 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
19820115 21719571 21347285 24845553 21282665 20798317 28004739 24508170 30249741 23537049 20566863 28648823 25401762 26823975 20075255 26334808 29403074 24495485 17510324 26483478 24438588 12364791 17994087 20966253 17550304 25348373 15632085 25244985 25830018 20920257 23761445 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
EMBL
BABH01003018
ODYU01002626
SOQ40376.1
NWSH01000070
PCG79983.1
KZ150017
+ More
PZC74949.1 GAIX01004589 JAA87971.1 RSAL01000014 RVE53249.1 KQ459596 KPI95461.1 AGBW02010351 OWR48656.1 GDQN01007201 JAT83853.1 KQ971318 EFA00436.1 KQ978430 KYM94009.1 GL888066 EGI68298.1 KQ976511 KYM82438.1 KQ981280 KYN43377.1 ADTU01024048 KQ982498 KYQ55672.1 KK852483 KDR22848.1 GECZ01029375 JAS40394.1 GL769036 EFZ10815.1 NEVH01012087 PNF30785.1 GECU01001198 JAT06509.1 GL437125 EFN70965.1 GEZM01062871 JAV69447.1 GL445584 EFN89316.1 GEDC01030425 GEDC01011992 JAS06873.1 JAS25306.1 KK107124 QOIP01000002 EZA58457.1 RLU25729.1 KZ288197 PBC33732.1 KQ414629 KOC67316.1 KQ980824 KYN12636.1 APGK01041389 KB740993 KB632256 ENN76082.1 ERL90701.1 DS235758 EEB16483.1 LJIG01022604 KRT79681.1 KQ434878 KZC09932.1 NNAY01000595 OXU27498.1 KQ435699 KOX80608.1 GBHO01040931 GBRD01002434 GDHC01012479 JAG02673.1 JAG63387.1 JAQ06150.1 GBGD01001389 JAC87500.1 NNAY01001008 OXU25509.1 GECL01001482 JAP04642.1 PYGN01000427 PSN46585.1 GGMS01001756 MBY70959.1 GFTR01006323 JAW10103.1 ABLF02031725 ACPB03014830 GAMC01001788 JAC04768.1 CVRI01000040 CRK94911.1 GFDF01000210 JAV13874.1 CCAG010011618 JXJN01000753 CH477228 EAT47065.1 GANO01003523 JAB56348.1 OUUW01000011 SPP86590.1 AXCN02000808 JXUM01098515 KQ564426 KXJ72305.1 JXUM01066977 KQ562426 KXJ75939.1 ATLV01023794 KE525346 KFB49570.1 GFDL01015309 JAV19736.1 AAAB01008964 CH902635 EDV33906.1 APCN01000778 EAA12281.4 CM000162 EDX02783.1 CH954180 EDV46645.1 GAKP01010338 JAC48614.1 CH379064 EDY73019.1 GGFK01010625 MBW43946.1 KRT07012.1 GDHF01032570 GDHF01026424 GDHF01017784 GDHF01007512 JAI19744.1 JAI25890.1 JAI34530.1 JAI44802.1 DS231838 EDS33867.1 AXCM01005173 AJVK01004229 CH933811 EDW06446.1 GBXI01010455 JAD03837.1 CH940651 EDW65174.2 ADMH02000622 ETN65574.1 CP012528 ALC49687.1 CH963851 EDW75392.2 CH480823 EDW56082.1 AE014298 AY061442 AAF48897.2 AAL28990.1
PZC74949.1 GAIX01004589 JAA87971.1 RSAL01000014 RVE53249.1 KQ459596 KPI95461.1 AGBW02010351 OWR48656.1 GDQN01007201 JAT83853.1 KQ971318 EFA00436.1 KQ978430 KYM94009.1 GL888066 EGI68298.1 KQ976511 KYM82438.1 KQ981280 KYN43377.1 ADTU01024048 KQ982498 KYQ55672.1 KK852483 KDR22848.1 GECZ01029375 JAS40394.1 GL769036 EFZ10815.1 NEVH01012087 PNF30785.1 GECU01001198 JAT06509.1 GL437125 EFN70965.1 GEZM01062871 JAV69447.1 GL445584 EFN89316.1 GEDC01030425 GEDC01011992 JAS06873.1 JAS25306.1 KK107124 QOIP01000002 EZA58457.1 RLU25729.1 KZ288197 PBC33732.1 KQ414629 KOC67316.1 KQ980824 KYN12636.1 APGK01041389 KB740993 KB632256 ENN76082.1 ERL90701.1 DS235758 EEB16483.1 LJIG01022604 KRT79681.1 KQ434878 KZC09932.1 NNAY01000595 OXU27498.1 KQ435699 KOX80608.1 GBHO01040931 GBRD01002434 GDHC01012479 JAG02673.1 JAG63387.1 JAQ06150.1 GBGD01001389 JAC87500.1 NNAY01001008 OXU25509.1 GECL01001482 JAP04642.1 PYGN01000427 PSN46585.1 GGMS01001756 MBY70959.1 GFTR01006323 JAW10103.1 ABLF02031725 ACPB03014830 GAMC01001788 JAC04768.1 CVRI01000040 CRK94911.1 GFDF01000210 JAV13874.1 CCAG010011618 JXJN01000753 CH477228 EAT47065.1 GANO01003523 JAB56348.1 OUUW01000011 SPP86590.1 AXCN02000808 JXUM01098515 KQ564426 KXJ72305.1 JXUM01066977 KQ562426 KXJ75939.1 ATLV01023794 KE525346 KFB49570.1 GFDL01015309 JAV19736.1 AAAB01008964 CH902635 EDV33906.1 APCN01000778 EAA12281.4 CM000162 EDX02783.1 CH954180 EDV46645.1 GAKP01010338 JAC48614.1 CH379064 EDY73019.1 GGFK01010625 MBW43946.1 KRT07012.1 GDHF01032570 GDHF01026424 GDHF01017784 GDHF01007512 JAI19744.1 JAI25890.1 JAI34530.1 JAI44802.1 DS231838 EDS33867.1 AXCM01005173 AJVK01004229 CH933811 EDW06446.1 GBXI01010455 JAD03837.1 CH940651 EDW65174.2 ADMH02000622 ETN65574.1 CP012528 ALC49687.1 CH963851 EDW75392.2 CH480823 EDW56082.1 AE014298 AY061442 AAF48897.2 AAL28990.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000007151
UP000007266
+ More
UP000078542 UP000007755 UP000078540 UP000078541 UP000005205 UP000075809 UP000027135 UP000235965 UP000000311 UP000008237 UP000053097 UP000279307 UP000242457 UP000053825 UP000078492 UP000005203 UP000019118 UP000030742 UP000009046 UP000076502 UP000215335 UP000053105 UP000002358 UP000245037 UP000007819 UP000015103 UP000183832 UP000092444 UP000078200 UP000092460 UP000091820 UP000008820 UP000268350 UP000075886 UP000069940 UP000249989 UP000030765 UP000007801 UP000075882 UP000075903 UP000076407 UP000075840 UP000192221 UP000007062 UP000002282 UP000075902 UP000075885 UP000008711 UP000075880 UP000075881 UP000075900 UP000001819 UP000075884 UP000076408 UP000002320 UP000069272 UP000075883 UP000092462 UP000075920 UP000009192 UP000008792 UP000000673 UP000092553 UP000095300 UP000095301 UP000007798 UP000001292 UP000000803
UP000078542 UP000007755 UP000078540 UP000078541 UP000005205 UP000075809 UP000027135 UP000235965 UP000000311 UP000008237 UP000053097 UP000279307 UP000242457 UP000053825 UP000078492 UP000005203 UP000019118 UP000030742 UP000009046 UP000076502 UP000215335 UP000053105 UP000002358 UP000245037 UP000007819 UP000015103 UP000183832 UP000092444 UP000078200 UP000092460 UP000091820 UP000008820 UP000268350 UP000075886 UP000069940 UP000249989 UP000030765 UP000007801 UP000075882 UP000075903 UP000076407 UP000075840 UP000192221 UP000007062 UP000002282 UP000075902 UP000075885 UP000008711 UP000075880 UP000075881 UP000075900 UP000001819 UP000075884 UP000076408 UP000002320 UP000069272 UP000075883 UP000092462 UP000075920 UP000009192 UP000008792 UP000000673 UP000092553 UP000095300 UP000095301 UP000007798 UP000001292 UP000000803
Interpro
SUPFAM
SSF54001
SSF54001
Gene 3D
ProteinModelPortal
H9JDS9
A0A2H1VHW3
A0A2A4K864
A0A2W1BIS2
S4P7I5
A0A437BST3
+ More
A0A194PPT9 A0A212F4J7 A0A1E1WA46 D6WET8 A0A151I745 F4WBM0 A0A195BD01 A0A195FSV0 A0A158NRL6 A0A151X5N3 A0A067RII4 A0A1B6ER25 E9J8N0 A0A2J7QQB8 A0A1B6K4Z3 E2A6G5 A0A1Y1LEJ8 E2B4N5 A0A1B6DHZ2 A0A026WQY7 A0A2A3ER45 A0A0L7R947 A0A195DII0 A0A088AFY9 N6UBQ8 E0VSX7 A0A0T6AXA6 A0A154PDK1 A0A232FAM9 A0A0N0U7V5 A0A0A9W861 K7J134 A0A069DU87 A0A232F4U6 A0A0V0G8Y6 A0A2P8YQR4 A0A2S2PZU1 A0A224XPL3 J9JPK8 T1HFN5 W8BZI9 A0A1J1I3J9 A0A1L8E5F3 A0A1B0FDZ7 A0A1A9VRS2 A0A1B0AN93 A0A1A9WSX4 Q17K27 U5ERX4 A0A3B0KE61 A0A182QCP5 A0A182GYZ3 A0A182GIQ4 A0A084WH76 A0A1Q3EWS9 A0A1S4GY37 B3MZV5 A0A182L0K3 A0A182ULV8 A0A182XFY0 A0A182IC66 A0A1W4U9X5 Q7Q4G1 B4PWG5 A0A182TTI1 A0A182PLR3 B3NUP8 A0A182IPX3 A0A182K645 A0A182RDR3 A0A034W1P3 B5DNL3 A0A2M4AT25 A0A182NTZ6 A0A0R3P1D0 A0A182YDU5 A0A0K8TZ59 B0W4M2 A0A182F3F9 A0A182MHL0 A0A1B0GMV9 A0A182W6F9 B4L5I7 A0A0A1WYF5 B4M2Z9 W5JS46 A0A0M4FAC4 A0A1I8P9J7 A0A1I8P9J3 A0A1I8MYQ0 A0A1I8MYP6 B4MTF3 B4I6Y4 Q9VWP1
A0A194PPT9 A0A212F4J7 A0A1E1WA46 D6WET8 A0A151I745 F4WBM0 A0A195BD01 A0A195FSV0 A0A158NRL6 A0A151X5N3 A0A067RII4 A0A1B6ER25 E9J8N0 A0A2J7QQB8 A0A1B6K4Z3 E2A6G5 A0A1Y1LEJ8 E2B4N5 A0A1B6DHZ2 A0A026WQY7 A0A2A3ER45 A0A0L7R947 A0A195DII0 A0A088AFY9 N6UBQ8 E0VSX7 A0A0T6AXA6 A0A154PDK1 A0A232FAM9 A0A0N0U7V5 A0A0A9W861 K7J134 A0A069DU87 A0A232F4U6 A0A0V0G8Y6 A0A2P8YQR4 A0A2S2PZU1 A0A224XPL3 J9JPK8 T1HFN5 W8BZI9 A0A1J1I3J9 A0A1L8E5F3 A0A1B0FDZ7 A0A1A9VRS2 A0A1B0AN93 A0A1A9WSX4 Q17K27 U5ERX4 A0A3B0KE61 A0A182QCP5 A0A182GYZ3 A0A182GIQ4 A0A084WH76 A0A1Q3EWS9 A0A1S4GY37 B3MZV5 A0A182L0K3 A0A182ULV8 A0A182XFY0 A0A182IC66 A0A1W4U9X5 Q7Q4G1 B4PWG5 A0A182TTI1 A0A182PLR3 B3NUP8 A0A182IPX3 A0A182K645 A0A182RDR3 A0A034W1P3 B5DNL3 A0A2M4AT25 A0A182NTZ6 A0A0R3P1D0 A0A182YDU5 A0A0K8TZ59 B0W4M2 A0A182F3F9 A0A182MHL0 A0A1B0GMV9 A0A182W6F9 B4L5I7 A0A0A1WYF5 B4M2Z9 W5JS46 A0A0M4FAC4 A0A1I8P9J7 A0A1I8P9J3 A0A1I8MYQ0 A0A1I8MYP6 B4MTF3 B4I6Y4 Q9VWP1
PDB
6AH0
E-value=4.6369e-180,
Score=1622
Ontologies
GO
PANTHER
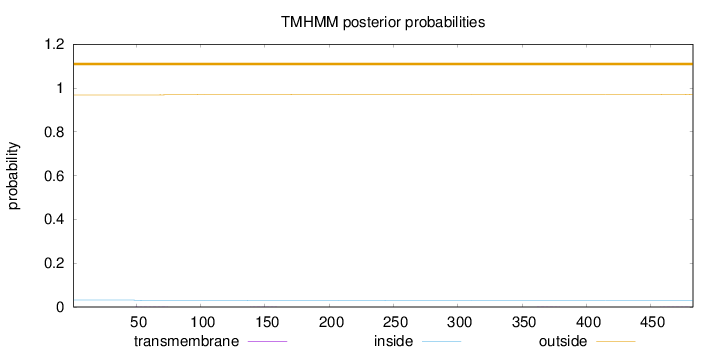
Topology
Length:
483
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0511300000000001
Exp number, first 60 AAs:
0.01191
Total prob of N-in:
0.03148
outside
1 - 483
Population Genetic Test Statistics
Pi
269.22487
Theta
189.772471
Tajima's D
1.316825
CLR
0.015041
CSRT
0.743662816859157
Interpretation
Uncertain
