Pre Gene Modal
BGIBMGA007905
Annotation
PREDICTED:_protein_purity_of_essence_[Bombyx_mori]
Full name
ATP-dependent DNA helicase
Location in the cell
Extracellular Reliability : 1.671
Sequence
CDS
ATGGCAGCTTCGGGAGGAAACATTGAATGGGCTAATACAGTGAAACCAATTTTAACAGCTCTCTACGGTCCTTTTTTCGATAAAAACGATGCTGTAGAGATAATTCAAGCAGTTATTAAGAGTGAGAATGAGTTCCAGAATCATGAAGACATGTATGATTTGTTCTACACATGTTTTGCATGTCTTTCTGCACATTTCATCAGTATCAATGCCAATAATTGTAAGTAA
Protein
MAASGGNIEWANTVKPILTALYGPFFDKNDAVEIIQAVIKSENEFQNHEDMYDLFYTCFACLSAHFISINANNCK
Summary
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Cofactor
Mg(2+)
Similarity
Belongs to the helicase family.
Uniprot
H9JEF8
A0A0N0PG51
A0A194PWF1
A0A2W1BJA7
A0A2A4K7U4
A0A1E1WBG5
+ More
A0A2H1VHR8 A0A212F4K1 A0A2J7QFT9 A0A2J7QFY7 A0A1B6LLA8 A0A2P8YQ54 A0A1B6EW24 A0A1B6J919 A0A0C9RH81 E0VLM7 A0A1B6CIJ6 A0A195C4Q7 A0A195BKW7 A0A158P3Z4 K7IMV9 A0A151JM14 F4WKS3 A0A232FE93 A0A026WJW0 E2AZ34 A0A3L8DAH1 A0A151JZ93 A0A1B6D105 A0A023F0S5 A0A069DYI5 E9J8Y0 V4CQY7 T1IY63 A0A423TEV8 A0A2R7WYZ6 A0A154PNA2 A0A0L7R551 H3BIS8 A0A1S3MIP3 A0A3Q3MHE0 A0A1S3MIE0 A0A1I8NYS2 A0A3N0YJM2 X1WEL5 A0A2R8QHT3 A0A0L0CMG0 Q4S817 H3D368 W5LRH0 W5MAW8 A0A2D0SIJ4 A0A2D0SIJ1 A0A2D0SJ52 A0A2D0SJP1 A0A2D0SII8 A0A2D0SII6 A0A2D0SIJ0 A0A2D0SJ45 A0A2D0SJN6 A0A060YPU2 A0A0P7XM31 A0A3P8QU03 A0A3P9CYU1 A0A3Q1C7C3 A0A3Q3WT24 A0A3B4VCM5 A0A3P8T8V8 A0A3Q3IQ95 A0A3Q0SZY7 A0A2I4ANV4 A0A3Q4G550 A0A3B4Z8V2 A0A3Q3BXQ1 A0A2I4ANR5 A0A3B5BHD0 A0A2U9C2Z2 A0A2I4ANT1 A0A2I4ANP7 A0A2I4ANQ8 A0A2I4ANU7 A0A2I4ANS6 A0A2I4ANP1 A0A2I4ANR1 A0A3P8YL95 A0A3P9J321 H2LGQ9 A0A3B3BF08 A0A3P9L4C4 A0A3S2PUY4 A0A3B4FTU0 A0A2P2IBR3 A0A0S7J4J0 A0A0S7J4E5 A0A0S7J7B6 A0A0S7J6B1 G3NT04 M4AWS3 A0A315W6U8
A0A2H1VHR8 A0A212F4K1 A0A2J7QFT9 A0A2J7QFY7 A0A1B6LLA8 A0A2P8YQ54 A0A1B6EW24 A0A1B6J919 A0A0C9RH81 E0VLM7 A0A1B6CIJ6 A0A195C4Q7 A0A195BKW7 A0A158P3Z4 K7IMV9 A0A151JM14 F4WKS3 A0A232FE93 A0A026WJW0 E2AZ34 A0A3L8DAH1 A0A151JZ93 A0A1B6D105 A0A023F0S5 A0A069DYI5 E9J8Y0 V4CQY7 T1IY63 A0A423TEV8 A0A2R7WYZ6 A0A154PNA2 A0A0L7R551 H3BIS8 A0A1S3MIP3 A0A3Q3MHE0 A0A1S3MIE0 A0A1I8NYS2 A0A3N0YJM2 X1WEL5 A0A2R8QHT3 A0A0L0CMG0 Q4S817 H3D368 W5LRH0 W5MAW8 A0A2D0SIJ4 A0A2D0SIJ1 A0A2D0SJ52 A0A2D0SJP1 A0A2D0SII8 A0A2D0SII6 A0A2D0SIJ0 A0A2D0SJ45 A0A2D0SJN6 A0A060YPU2 A0A0P7XM31 A0A3P8QU03 A0A3P9CYU1 A0A3Q1C7C3 A0A3Q3WT24 A0A3B4VCM5 A0A3P8T8V8 A0A3Q3IQ95 A0A3Q0SZY7 A0A2I4ANV4 A0A3Q4G550 A0A3B4Z8V2 A0A3Q3BXQ1 A0A2I4ANR5 A0A3B5BHD0 A0A2U9C2Z2 A0A2I4ANT1 A0A2I4ANP7 A0A2I4ANQ8 A0A2I4ANU7 A0A2I4ANS6 A0A2I4ANP1 A0A2I4ANR1 A0A3P8YL95 A0A3P9J321 H2LGQ9 A0A3B3BF08 A0A3P9L4C4 A0A3S2PUY4 A0A3B4FTU0 A0A2P2IBR3 A0A0S7J4J0 A0A0S7J4E5 A0A0S7J7B6 A0A0S7J6B1 G3NT04 M4AWS3 A0A315W6U8
EC Number
3.6.4.12
Pubmed
EMBL
BABH01003018
BABH01003019
BABH01003020
BABH01003021
BABH01003022
LADJ01061705
+ More
KPJ21622.1 KQ459596 KPI95460.1 KZ150017 PZC74948.1 NWSH01000070 PCG79984.1 GDQN01006777 JAT84277.1 ODYU01002626 SOQ40377.1 AGBW02010351 OWR48657.1 NEVH01014842 PNF27454.1 PNF27453.1 GEBQ01015462 JAT24515.1 PYGN01000438 PSN46385.1 GECZ01027630 JAS42139.1 GECU01012071 JAS95635.1 GBYB01012567 JAG82334.1 DS235277 EEB14283.1 GEDC01024029 JAS13269.1 KQ978317 KYM95161.1 KQ976453 KYM85360.1 ADTU01008643 ADTU01008644 ADTU01008645 KQ978949 KYN27374.1 GL888206 EGI65165.1 NNAY01000352 OXU28982.1 KK107167 EZA56295.1 GL444078 EFN61308.1 QOIP01000010 RLU17495.1 KQ981427 KYN41757.1 GEDC01017919 JAS19379.1 GBBI01004128 JAC14584.1 GBGD01000002 JAC88887.1 GL769162 EFZ10719.1 KB199651 ESP04880.1 JH431671 QCYY01001817 ROT75024.1 KK856060 PTY24747.1 KQ434998 KZC13346.1 KQ414654 KOC65881.1 AFYH01001000 AFYH01001001 AFYH01001002 AFYH01001003 AFYH01001004 AFYH01001005 AFYH01001006 AFYH01001007 AFYH01001008 AFYH01001009 RJVU01040995 ROL46110.1 CT027552 JRES01000186 KNC33485.1 CAAE01014710 CAG03215.1 AHAT01009767 AHAT01009768 AHAT01009769 AHAT01009770 AHAT01009771 AHAT01009772 FR915544 CDQ93597.1 JARO02000639 KPP77868.1 CP026253 AWP09422.1 CM012443 RVE70356.1 IACF01005865 LAB71448.1 GBYX01248845 GBYX01248812 JAO60183.1 GBYX01248896 GBYX01248890 GBYX01248881 GBYX01248874 GBYX01248866 GBYX01248862 GBYX01248859 GBYX01248858 GBYX01248856 GBYX01248855 GBYX01248853 GBYX01248849 GBYX01248846 GBYX01248834 GBYX01248832 GBYX01248827 GBYX01248795 GBYX01248791 JAO60145.1 GBYX01248888 GBYX01248809 JAO60186.1 GBYX01248884 GBYX01248878 JAO60121.1 NHOQ01000318 PWA31070.1
KPJ21622.1 KQ459596 KPI95460.1 KZ150017 PZC74948.1 NWSH01000070 PCG79984.1 GDQN01006777 JAT84277.1 ODYU01002626 SOQ40377.1 AGBW02010351 OWR48657.1 NEVH01014842 PNF27454.1 PNF27453.1 GEBQ01015462 JAT24515.1 PYGN01000438 PSN46385.1 GECZ01027630 JAS42139.1 GECU01012071 JAS95635.1 GBYB01012567 JAG82334.1 DS235277 EEB14283.1 GEDC01024029 JAS13269.1 KQ978317 KYM95161.1 KQ976453 KYM85360.1 ADTU01008643 ADTU01008644 ADTU01008645 KQ978949 KYN27374.1 GL888206 EGI65165.1 NNAY01000352 OXU28982.1 KK107167 EZA56295.1 GL444078 EFN61308.1 QOIP01000010 RLU17495.1 KQ981427 KYN41757.1 GEDC01017919 JAS19379.1 GBBI01004128 JAC14584.1 GBGD01000002 JAC88887.1 GL769162 EFZ10719.1 KB199651 ESP04880.1 JH431671 QCYY01001817 ROT75024.1 KK856060 PTY24747.1 KQ434998 KZC13346.1 KQ414654 KOC65881.1 AFYH01001000 AFYH01001001 AFYH01001002 AFYH01001003 AFYH01001004 AFYH01001005 AFYH01001006 AFYH01001007 AFYH01001008 AFYH01001009 RJVU01040995 ROL46110.1 CT027552 JRES01000186 KNC33485.1 CAAE01014710 CAG03215.1 AHAT01009767 AHAT01009768 AHAT01009769 AHAT01009770 AHAT01009771 AHAT01009772 FR915544 CDQ93597.1 JARO02000639 KPP77868.1 CP026253 AWP09422.1 CM012443 RVE70356.1 IACF01005865 LAB71448.1 GBYX01248845 GBYX01248812 JAO60183.1 GBYX01248896 GBYX01248890 GBYX01248881 GBYX01248874 GBYX01248866 GBYX01248862 GBYX01248859 GBYX01248858 GBYX01248856 GBYX01248855 GBYX01248853 GBYX01248849 GBYX01248846 GBYX01248834 GBYX01248832 GBYX01248827 GBYX01248795 GBYX01248791 JAO60145.1 GBYX01248888 GBYX01248809 JAO60186.1 GBYX01248884 GBYX01248878 JAO60121.1 NHOQ01000318 PWA31070.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000007151
UP000235965
+ More
UP000245037 UP000009046 UP000078542 UP000078540 UP000005205 UP000002358 UP000078492 UP000007755 UP000215335 UP000053097 UP000000311 UP000279307 UP000078541 UP000030746 UP000283509 UP000076502 UP000053825 UP000008672 UP000087266 UP000261640 UP000095300 UP000000437 UP000037069 UP000007303 UP000018467 UP000018468 UP000221080 UP000193380 UP000034805 UP000265100 UP000265160 UP000257160 UP000261620 UP000261420 UP000265080 UP000261600 UP000261340 UP000192220 UP000261580 UP000261360 UP000264840 UP000261400 UP000246464 UP000265140 UP000265200 UP000001038 UP000261560 UP000265180 UP000261460 UP000007635 UP000002852
UP000245037 UP000009046 UP000078542 UP000078540 UP000005205 UP000002358 UP000078492 UP000007755 UP000215335 UP000053097 UP000000311 UP000279307 UP000078541 UP000030746 UP000283509 UP000076502 UP000053825 UP000008672 UP000087266 UP000261640 UP000095300 UP000000437 UP000037069 UP000007303 UP000018467 UP000018468 UP000221080 UP000193380 UP000034805 UP000265100 UP000265160 UP000257160 UP000261620 UP000261420 UP000265080 UP000261600 UP000261340 UP000192220 UP000261580 UP000261360 UP000264840 UP000261400 UP000246464 UP000265140 UP000265200 UP000001038 UP000261560 UP000265180 UP000261460 UP000007635 UP000002852
PRIDE
Interpro
ProteinModelPortal
H9JEF8
A0A0N0PG51
A0A194PWF1
A0A2W1BJA7
A0A2A4K7U4
A0A1E1WBG5
+ More
A0A2H1VHR8 A0A212F4K1 A0A2J7QFT9 A0A2J7QFY7 A0A1B6LLA8 A0A2P8YQ54 A0A1B6EW24 A0A1B6J919 A0A0C9RH81 E0VLM7 A0A1B6CIJ6 A0A195C4Q7 A0A195BKW7 A0A158P3Z4 K7IMV9 A0A151JM14 F4WKS3 A0A232FE93 A0A026WJW0 E2AZ34 A0A3L8DAH1 A0A151JZ93 A0A1B6D105 A0A023F0S5 A0A069DYI5 E9J8Y0 V4CQY7 T1IY63 A0A423TEV8 A0A2R7WYZ6 A0A154PNA2 A0A0L7R551 H3BIS8 A0A1S3MIP3 A0A3Q3MHE0 A0A1S3MIE0 A0A1I8NYS2 A0A3N0YJM2 X1WEL5 A0A2R8QHT3 A0A0L0CMG0 Q4S817 H3D368 W5LRH0 W5MAW8 A0A2D0SIJ4 A0A2D0SIJ1 A0A2D0SJ52 A0A2D0SJP1 A0A2D0SII8 A0A2D0SII6 A0A2D0SIJ0 A0A2D0SJ45 A0A2D0SJN6 A0A060YPU2 A0A0P7XM31 A0A3P8QU03 A0A3P9CYU1 A0A3Q1C7C3 A0A3Q3WT24 A0A3B4VCM5 A0A3P8T8V8 A0A3Q3IQ95 A0A3Q0SZY7 A0A2I4ANV4 A0A3Q4G550 A0A3B4Z8V2 A0A3Q3BXQ1 A0A2I4ANR5 A0A3B5BHD0 A0A2U9C2Z2 A0A2I4ANT1 A0A2I4ANP7 A0A2I4ANQ8 A0A2I4ANU7 A0A2I4ANS6 A0A2I4ANP1 A0A2I4ANR1 A0A3P8YL95 A0A3P9J321 H2LGQ9 A0A3B3BF08 A0A3P9L4C4 A0A3S2PUY4 A0A3B4FTU0 A0A2P2IBR3 A0A0S7J4J0 A0A0S7J4E5 A0A0S7J7B6 A0A0S7J6B1 G3NT04 M4AWS3 A0A315W6U8
A0A2H1VHR8 A0A212F4K1 A0A2J7QFT9 A0A2J7QFY7 A0A1B6LLA8 A0A2P8YQ54 A0A1B6EW24 A0A1B6J919 A0A0C9RH81 E0VLM7 A0A1B6CIJ6 A0A195C4Q7 A0A195BKW7 A0A158P3Z4 K7IMV9 A0A151JM14 F4WKS3 A0A232FE93 A0A026WJW0 E2AZ34 A0A3L8DAH1 A0A151JZ93 A0A1B6D105 A0A023F0S5 A0A069DYI5 E9J8Y0 V4CQY7 T1IY63 A0A423TEV8 A0A2R7WYZ6 A0A154PNA2 A0A0L7R551 H3BIS8 A0A1S3MIP3 A0A3Q3MHE0 A0A1S3MIE0 A0A1I8NYS2 A0A3N0YJM2 X1WEL5 A0A2R8QHT3 A0A0L0CMG0 Q4S817 H3D368 W5LRH0 W5MAW8 A0A2D0SIJ4 A0A2D0SIJ1 A0A2D0SJ52 A0A2D0SJP1 A0A2D0SII8 A0A2D0SII6 A0A2D0SIJ0 A0A2D0SJ45 A0A2D0SJN6 A0A060YPU2 A0A0P7XM31 A0A3P8QU03 A0A3P9CYU1 A0A3Q1C7C3 A0A3Q3WT24 A0A3B4VCM5 A0A3P8T8V8 A0A3Q3IQ95 A0A3Q0SZY7 A0A2I4ANV4 A0A3Q4G550 A0A3B4Z8V2 A0A3Q3BXQ1 A0A2I4ANR5 A0A3B5BHD0 A0A2U9C2Z2 A0A2I4ANT1 A0A2I4ANP7 A0A2I4ANQ8 A0A2I4ANU7 A0A2I4ANS6 A0A2I4ANP1 A0A2I4ANR1 A0A3P8YL95 A0A3P9J321 H2LGQ9 A0A3B3BF08 A0A3P9L4C4 A0A3S2PUY4 A0A3B4FTU0 A0A2P2IBR3 A0A0S7J4J0 A0A0S7J4E5 A0A0S7J7B6 A0A0S7J6B1 G3NT04 M4AWS3 A0A315W6U8
Ontologies
KEGG
GO
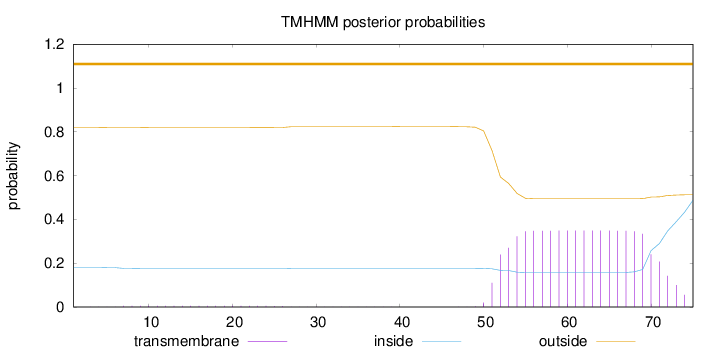
Topology
Length:
75
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
7.02303
Exp number, first 60 AAs:
3.1615
Total prob of N-in:
0.18061
outside
1 - 75
Population Genetic Test Statistics
Pi
213.246739
Theta
160.830144
Tajima's D
1.024882
CLR
0.000092
CSRT
0.668116594170291
Interpretation
Uncertain
