Gene
KWMTBOMO08738
Pre Gene Modal
BGIBMGA007907
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_SET_and_MYND_domain-containing_protein_4_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.272
Sequence
CDS
ATGACCGAAGAAAACGAAGGTTTCTTTAAGAAGTTTCACGAATCGATCAACAACGAATTAGACGAAGTCACAAAAAACAATTTCGCCAATTTAGAATCGAACTCTAAGCGCGTTTCGTATCTGTGCAGTCTCCCGATTATAAAAAAGTACGATATCACCGATGATGTTGTGAAATGTAAAACGGGTGGCGAGTTTCCTGTAAACAAATCTTTAAGCAAAGCTCTACAATTAAAGGACGCGGGAAACCAATGGGTCCAAAAGGGTGATTGGGTGAAGGCTTTGGTATTTTACAGTCAAAGTATGATGCATATGCCAGAAAAGGAAACGGAAGAGTTGTCAATTGTTTTCGCCAATCGATCGGCAGCTCTGAACCATTTGCAGCAATACGAGGAAGCCCTAGCCGATATAAAGAAATGTTTGTCCCTCGGATATCCTCGGCATCTCAGATACAAAGTTTACGAAAGAAAAGCACGTAGTTTGTTGGCGCTGAAAAGGAACCGGGAATCAATTGTTGCATTCCAAGACACTATAAGTGCTTTAGATGAGGCAACAAAACTTGAAAAGGAGAAAAGAACCAAAATGCGGACAGATGCTAAGATAATGTTGGAAATTTTGAACAAGGGACTAGTTCTCGCTGGGAATCCAAGCGATCCAGAACCTTTAAATAAGATTCCTCCAAAACCGAAATTAGCAGGAAAGCATAACTCCAAGTTCCCGGCGGCTTCTGAAGCGATAACAATAGATACTGACGACGTTAAAGGAAGATACGCTACTGCAACAAGGGATATCAGTGCTGGAGAAGTTTTGCTTATTGAGAAAGCGCACAGTGGAGTTCTTTTGGCTGAATTCTCGAAGACACATTGTCAAAATTGCTTCCTTAAGTGTCCTATATCACTACCGTGTCCCAAGTGTCCGAACGTAATATTCTGCAGCGACAAATGCCTAGAAACCGCTTTAAAATCATATCACGGTTACGAATGTCAAATACTTCCATTGATATGGAAATCTGGATGTTCTATCACATGCCAAATCGCTTTGAGAATGATCACGCAAAACAAAAAGGATTACTTTTTGAGCATCAGCAATGATTTGAAGCCCCAGTTGGGTGCATACAAGACGGAAGACTACAGAAACATTTACAATTTAACAACTCATGAAGAAAAAAGATGCAAGCAAGACTTTTTGCACAGAACAGAGATGGTAGTATTCCTGATTAAGGTCTTAGAATTGACCGGTTATTTTGACGGAAAACCAAGAACGAAGCCGATAGAAATGGTGGAACTGAATTCAATGGCGGTTAACGAAAAATACCAGGGTGACGTAGAATTGATAGGTGGCTACTTGTTGAGGAATCTTCAGGTTCTACAATTCAATGCGCACGAAGTGTTTGAACTTCAAGTACCTAAGCCGAAAGTTGATAAGAATATTATAAAACATGACGGAAAATCAGTGTTTTTAGCCGGAGCCGTTTTCCCGACACTCGCCCTATTCAATCACTCCTGTGAACCAGGCGTTGTAAGATATTTCTATGGCTCACACATCGTAGTTCGTGCCGTCAAAAATATCAAAAAAGGTGAGGAAGTCTGCGAAAACTACGGTCCAATATTCACGACCGTGCCCAAGGAGAAGAGACAAATGGAACTCAAAGACCGGTACTGGTTTGACTGTGAATGCATACCGTGCAAGGAAAACTGGCCTTTGTACGATGAGATGACCGAGAATTACATGAGATTCAAGTGCGATTCCGACAATCCTTGCCCGAACGTGGTGGCTGTTCCGTACGACTGCACCGAATTCATGATCCAGTGTGGTTTATGCCAGCAGTACACCAACATACTGAAGGGACTAAAATCGCTCCAGAACCAGAAATCGAAAATCTTACTAAAATTAGCACCAGTTCAAAGTTTGTCCGGTATAATATATGGAAATGGTGTCTTCTCGAAAATTACAAAGCAAACAACTCTATACAAAGAACAAAACTTTTAA
Protein
MTEENEGFFKKFHESINNELDEVTKNNFANLESNSKRVSYLCSLPIIKKYDITDDVVKCKTGGEFPVNKSLSKALQLKDAGNQWVQKGDWVKALVFYSQSMMHMPEKETEELSIVFANRSAALNHLQQYEEALADIKKCLSLGYPRHLRYKVYERKARSLLALKRNRESIVAFQDTISALDEATKLEKEKRTKMRTDAKIMLEILNKGLVLAGNPSDPEPLNKIPPKPKLAGKHNSKFPAASEAITIDTDDVKGRYATATRDISAGEVLLIEKAHSGVLLAEFSKTHCQNCFLKCPISLPCPKCPNVIFCSDKCLETALKSYHGYECQILPLIWKSGCSITCQIALRMITQNKKDYFLSISNDLKPQLGAYKTEDYRNIYNLTTHEEKRCKQDFLHRTEMVVFLIKVLELTGYFDGKPRTKPIEMVELNSMAVNEKYQGDVELIGGYLLRNLQVLQFNAHEVFELQVPKPKVDKNIIKHDGKSVFLAGAVFPTLALFNHSCEPGVVRYFYGSHIVVRAVKNIKKGEEVCENYGPIFTTVPKEKRQMELKDRYWFDCECIPCKENWPLYDEMTENYMRFKCDSDNPCPNVVAVPYDCTEFMIQCGLCQQYTNILKGLKSLQNQKSKILLKLAPVQSLSGIIYGNGVFSKITKQTTLYKEQNF
Summary
Uniprot
H9JEG0
A0A2H1V057
A0A2A4JB22
A0A3S2LT25
A0A194QM83
A0A194PPT4
+ More
A0A212ERL4 A0A0L7KXA3 A0A2W1B565 A0A1B6F3Y7 A0A146LER3 A0A0K8T434 A0A0A9YT27 A0A1B6J8P5 A0A023F0G4 A0A1B6CWC9 T1I1H4 A0A2J7RM43 A0A1B0CSA0 Q0IG30 A0A067RIA7 B0X7W3 E0VHL0 A0A0A1XQ85 A0A034VBD7 A0A1S4F4X7 W8BM99 A0A0K8VG01 A0A182QSN0 A0A182Y978 A0A182H4D0 W5JSF9 A0A182TQX6 Q7PWV2 A0A182UST0 A0A1W4UGC5 A0A2C9GRY7 A0A3B0JEX9 A0A182VU51 A0A182T3J0 A0A182LXV7 B3MFB7 A0A087ZR37 A0A182JZN0 A0A1A9YPG3 A0A084VYF9 A0A1B0BI81 B3NSG0 A0A195CR60 A0A0J9TZ25 B4J8Z5 A1Z8L3 A0A1B0G1K5 B4HNH7 A0A3L8E364 A0A0R1DWA2 A0A1I8NI76 A0A182R4A7 A0A1I8QEF5 A0A1B0A9A5 B4LQ48 A0A310SJH7 A0A026WG26 B4P647 Q28YA2 A0A1A9UIY6 A0A182KNG7 A0A0J7L0U2 A0A2A3E941 A0A182XDH9 E9IAB4 B4GCW8 A0A154PFP6 A0A0L0CN66 A0A151XIR9 A0A0N0U786 A0A182NUN6 A0A182JHR4 B4KR24 A0A1B6HSN7 J9JK42 A0A0M3QUU8 B4MQJ4 A0A224XAZ1 A0A195E0C5 E2B789 A0A158NES4 A0A195AVV3 Q95RZ8 B4QBL1 A0A0Q9WCE6 A0A195FKV7 E2A170 A0A0Q9XMD3 A0A0P4W687 K7JCG6 F4WEE6 A0A182F1Y5 A0A1B6L459 A0A0P4W8Y7
A0A212ERL4 A0A0L7KXA3 A0A2W1B565 A0A1B6F3Y7 A0A146LER3 A0A0K8T434 A0A0A9YT27 A0A1B6J8P5 A0A023F0G4 A0A1B6CWC9 T1I1H4 A0A2J7RM43 A0A1B0CSA0 Q0IG30 A0A067RIA7 B0X7W3 E0VHL0 A0A0A1XQ85 A0A034VBD7 A0A1S4F4X7 W8BM99 A0A0K8VG01 A0A182QSN0 A0A182Y978 A0A182H4D0 W5JSF9 A0A182TQX6 Q7PWV2 A0A182UST0 A0A1W4UGC5 A0A2C9GRY7 A0A3B0JEX9 A0A182VU51 A0A182T3J0 A0A182LXV7 B3MFB7 A0A087ZR37 A0A182JZN0 A0A1A9YPG3 A0A084VYF9 A0A1B0BI81 B3NSG0 A0A195CR60 A0A0J9TZ25 B4J8Z5 A1Z8L3 A0A1B0G1K5 B4HNH7 A0A3L8E364 A0A0R1DWA2 A0A1I8NI76 A0A182R4A7 A0A1I8QEF5 A0A1B0A9A5 B4LQ48 A0A310SJH7 A0A026WG26 B4P647 Q28YA2 A0A1A9UIY6 A0A182KNG7 A0A0J7L0U2 A0A2A3E941 A0A182XDH9 E9IAB4 B4GCW8 A0A154PFP6 A0A0L0CN66 A0A151XIR9 A0A0N0U786 A0A182NUN6 A0A182JHR4 B4KR24 A0A1B6HSN7 J9JK42 A0A0M3QUU8 B4MQJ4 A0A224XAZ1 A0A195E0C5 E2B789 A0A158NES4 A0A195AVV3 Q95RZ8 B4QBL1 A0A0Q9WCE6 A0A195FKV7 E2A170 A0A0Q9XMD3 A0A0P4W687 K7JCG6 F4WEE6 A0A182F1Y5 A0A1B6L459 A0A0P4W8Y7
Pubmed
19121390
26354079
22118469
26227816
28756777
26823975
+ More
25401762 25474469 17510324 24845553 20566863 25830018 25348373 24495485 25244985 26483478 20920257 23761445 12364791 14747013 17210077 17994087 24438588 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 30249741 17550304 25315136 24508170 15632085 20966253 21282665 26108605 20798317 21347285 20075255 21719571
25401762 25474469 17510324 24845553 20566863 25830018 25348373 24495485 25244985 26483478 20920257 23761445 12364791 14747013 17210077 17994087 24438588 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 30249741 17550304 25315136 24508170 15632085 20966253 21282665 26108605 20798317 21347285 20075255 21719571
EMBL
BABH01003039
ODYU01000077
SOQ34225.1
NWSH01002176
PCG68966.1
RSAL01000014
+ More
RVE53246.1 KQ461195 KPJ06648.1 KQ459596 KPI95456.1 AGBW02013012 OWR44094.1 JTDY01004862 KOB67664.1 KZ150318 PZC71432.1 GECZ01024885 JAS44884.1 GDHC01012550 JAQ06079.1 GBRD01005670 JAG60151.1 GBHO01008823 JAG34781.1 GECU01012149 JAS95557.1 GBBI01003755 JAC14957.1 GEDC01019561 JAS17737.1 ACPB03005171 NEVH01002577 PNF41905.1 AJWK01025809 CH477274 EAT45183.1 KK852460 KDR23527.1 DS232465 EDS42182.1 DS235171 EEB12896.1 GBXI01001257 JAD13035.1 GAKP01020096 GAKP01020094 JAC38858.1 GAMC01006634 GAMC01006633 JAB99922.1 GDHF01014460 JAI37854.1 AXCN02000734 JXUM01001256 JXUM01001257 JXUM01001258 JXUM01001259 ADMH02000599 ETN65694.1 AAAB01008984 EAA14823.4 APCN01003350 OUUW01000001 SPP73850.1 AXCM01000044 CH902619 EDV35591.1 ATLV01018364 KE525231 KFB43003.1 JXJN01014861 CH954179 EDV56462.1 KQ977381 KYN03191.1 CM002911 KMY93000.1 CH916367 EDW01344.1 AE013599 AAF58645.2 CCAG010020139 CH480816 EDW47413.1 QOIP01000001 RLU27144.1 CM000158 KRJ99520.1 CH940648 EDW61333.1 KQ759804 OAD62811.1 KK107260 EZA53989.1 EDW90922.2 CM000071 EAL26063.1 LBMM01001562 KMQ96084.1 KZ288344 PBC27709.1 GL761997 EFZ22489.1 CH479181 EDW32531.1 KQ434893 KZC10665.1 JRES01000160 KNC33731.1 KQ982080 KYQ60227.1 KQ435709 KOX79971.1 CH933808 EDW10380.1 GECU01030008 JAS77698.1 ABLF02026503 CP012524 ALC41289.1 CH963849 EDW74383.1 GFTR01006860 JAW09566.1 KQ979999 KYN18329.1 GL446149 EFN88450.1 ADTU01013628 KQ976736 KYM76099.1 AY061023 AAL28571.1 CM000362 EDX06628.1 KRF79927.1 KQ981490 KYN41300.1 GL435707 EFN72837.1 KRG05208.1 GDRN01080085 JAI62291.1 AAZX01003663 GL888103 EGI67433.1 GEBQ01021455 JAT18522.1 GDRN01080086 GDRN01080084 GDRN01080083 JAI62293.1
RVE53246.1 KQ461195 KPJ06648.1 KQ459596 KPI95456.1 AGBW02013012 OWR44094.1 JTDY01004862 KOB67664.1 KZ150318 PZC71432.1 GECZ01024885 JAS44884.1 GDHC01012550 JAQ06079.1 GBRD01005670 JAG60151.1 GBHO01008823 JAG34781.1 GECU01012149 JAS95557.1 GBBI01003755 JAC14957.1 GEDC01019561 JAS17737.1 ACPB03005171 NEVH01002577 PNF41905.1 AJWK01025809 CH477274 EAT45183.1 KK852460 KDR23527.1 DS232465 EDS42182.1 DS235171 EEB12896.1 GBXI01001257 JAD13035.1 GAKP01020096 GAKP01020094 JAC38858.1 GAMC01006634 GAMC01006633 JAB99922.1 GDHF01014460 JAI37854.1 AXCN02000734 JXUM01001256 JXUM01001257 JXUM01001258 JXUM01001259 ADMH02000599 ETN65694.1 AAAB01008984 EAA14823.4 APCN01003350 OUUW01000001 SPP73850.1 AXCM01000044 CH902619 EDV35591.1 ATLV01018364 KE525231 KFB43003.1 JXJN01014861 CH954179 EDV56462.1 KQ977381 KYN03191.1 CM002911 KMY93000.1 CH916367 EDW01344.1 AE013599 AAF58645.2 CCAG010020139 CH480816 EDW47413.1 QOIP01000001 RLU27144.1 CM000158 KRJ99520.1 CH940648 EDW61333.1 KQ759804 OAD62811.1 KK107260 EZA53989.1 EDW90922.2 CM000071 EAL26063.1 LBMM01001562 KMQ96084.1 KZ288344 PBC27709.1 GL761997 EFZ22489.1 CH479181 EDW32531.1 KQ434893 KZC10665.1 JRES01000160 KNC33731.1 KQ982080 KYQ60227.1 KQ435709 KOX79971.1 CH933808 EDW10380.1 GECU01030008 JAS77698.1 ABLF02026503 CP012524 ALC41289.1 CH963849 EDW74383.1 GFTR01006860 JAW09566.1 KQ979999 KYN18329.1 GL446149 EFN88450.1 ADTU01013628 KQ976736 KYM76099.1 AY061023 AAL28571.1 CM000362 EDX06628.1 KRF79927.1 KQ981490 KYN41300.1 GL435707 EFN72837.1 KRG05208.1 GDRN01080085 JAI62291.1 AAZX01003663 GL888103 EGI67433.1 GEBQ01021455 JAT18522.1 GDRN01080086 GDRN01080084 GDRN01080083 JAI62293.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000053268
UP000007151
+ More
UP000037510 UP000015103 UP000235965 UP000092461 UP000008820 UP000027135 UP000002320 UP000009046 UP000075886 UP000076408 UP000069940 UP000000673 UP000075902 UP000007062 UP000075903 UP000192221 UP000075840 UP000268350 UP000075920 UP000075901 UP000075883 UP000007801 UP000005203 UP000075881 UP000092443 UP000030765 UP000092460 UP000008711 UP000078542 UP000001070 UP000000803 UP000092444 UP000001292 UP000279307 UP000002282 UP000095301 UP000075900 UP000095300 UP000092445 UP000008792 UP000053097 UP000001819 UP000078200 UP000075882 UP000036403 UP000242457 UP000076407 UP000008744 UP000076502 UP000037069 UP000075809 UP000053105 UP000075884 UP000075880 UP000009192 UP000007819 UP000092553 UP000007798 UP000078492 UP000008237 UP000005205 UP000078540 UP000000304 UP000078541 UP000000311 UP000002358 UP000007755 UP000069272
UP000037510 UP000015103 UP000235965 UP000092461 UP000008820 UP000027135 UP000002320 UP000009046 UP000075886 UP000076408 UP000069940 UP000000673 UP000075902 UP000007062 UP000075903 UP000192221 UP000075840 UP000268350 UP000075920 UP000075901 UP000075883 UP000007801 UP000005203 UP000075881 UP000092443 UP000030765 UP000092460 UP000008711 UP000078542 UP000001070 UP000000803 UP000092444 UP000001292 UP000279307 UP000002282 UP000095301 UP000075900 UP000095300 UP000092445 UP000008792 UP000053097 UP000001819 UP000078200 UP000075882 UP000036403 UP000242457 UP000076407 UP000008744 UP000076502 UP000037069 UP000075809 UP000053105 UP000075884 UP000075880 UP000009192 UP000007819 UP000092553 UP000007798 UP000078492 UP000008237 UP000005205 UP000078540 UP000000304 UP000078541 UP000000311 UP000002358 UP000007755 UP000069272
Interpro
Gene 3D
ProteinModelPortal
H9JEG0
A0A2H1V057
A0A2A4JB22
A0A3S2LT25
A0A194QM83
A0A194PPT4
+ More
A0A212ERL4 A0A0L7KXA3 A0A2W1B565 A0A1B6F3Y7 A0A146LER3 A0A0K8T434 A0A0A9YT27 A0A1B6J8P5 A0A023F0G4 A0A1B6CWC9 T1I1H4 A0A2J7RM43 A0A1B0CSA0 Q0IG30 A0A067RIA7 B0X7W3 E0VHL0 A0A0A1XQ85 A0A034VBD7 A0A1S4F4X7 W8BM99 A0A0K8VG01 A0A182QSN0 A0A182Y978 A0A182H4D0 W5JSF9 A0A182TQX6 Q7PWV2 A0A182UST0 A0A1W4UGC5 A0A2C9GRY7 A0A3B0JEX9 A0A182VU51 A0A182T3J0 A0A182LXV7 B3MFB7 A0A087ZR37 A0A182JZN0 A0A1A9YPG3 A0A084VYF9 A0A1B0BI81 B3NSG0 A0A195CR60 A0A0J9TZ25 B4J8Z5 A1Z8L3 A0A1B0G1K5 B4HNH7 A0A3L8E364 A0A0R1DWA2 A0A1I8NI76 A0A182R4A7 A0A1I8QEF5 A0A1B0A9A5 B4LQ48 A0A310SJH7 A0A026WG26 B4P647 Q28YA2 A0A1A9UIY6 A0A182KNG7 A0A0J7L0U2 A0A2A3E941 A0A182XDH9 E9IAB4 B4GCW8 A0A154PFP6 A0A0L0CN66 A0A151XIR9 A0A0N0U786 A0A182NUN6 A0A182JHR4 B4KR24 A0A1B6HSN7 J9JK42 A0A0M3QUU8 B4MQJ4 A0A224XAZ1 A0A195E0C5 E2B789 A0A158NES4 A0A195AVV3 Q95RZ8 B4QBL1 A0A0Q9WCE6 A0A195FKV7 E2A170 A0A0Q9XMD3 A0A0P4W687 K7JCG6 F4WEE6 A0A182F1Y5 A0A1B6L459 A0A0P4W8Y7
A0A212ERL4 A0A0L7KXA3 A0A2W1B565 A0A1B6F3Y7 A0A146LER3 A0A0K8T434 A0A0A9YT27 A0A1B6J8P5 A0A023F0G4 A0A1B6CWC9 T1I1H4 A0A2J7RM43 A0A1B0CSA0 Q0IG30 A0A067RIA7 B0X7W3 E0VHL0 A0A0A1XQ85 A0A034VBD7 A0A1S4F4X7 W8BM99 A0A0K8VG01 A0A182QSN0 A0A182Y978 A0A182H4D0 W5JSF9 A0A182TQX6 Q7PWV2 A0A182UST0 A0A1W4UGC5 A0A2C9GRY7 A0A3B0JEX9 A0A182VU51 A0A182T3J0 A0A182LXV7 B3MFB7 A0A087ZR37 A0A182JZN0 A0A1A9YPG3 A0A084VYF9 A0A1B0BI81 B3NSG0 A0A195CR60 A0A0J9TZ25 B4J8Z5 A1Z8L3 A0A1B0G1K5 B4HNH7 A0A3L8E364 A0A0R1DWA2 A0A1I8NI76 A0A182R4A7 A0A1I8QEF5 A0A1B0A9A5 B4LQ48 A0A310SJH7 A0A026WG26 B4P647 Q28YA2 A0A1A9UIY6 A0A182KNG7 A0A0J7L0U2 A0A2A3E941 A0A182XDH9 E9IAB4 B4GCW8 A0A154PFP6 A0A0L0CN66 A0A151XIR9 A0A0N0U786 A0A182NUN6 A0A182JHR4 B4KR24 A0A1B6HSN7 J9JK42 A0A0M3QUU8 B4MQJ4 A0A224XAZ1 A0A195E0C5 E2B789 A0A158NES4 A0A195AVV3 Q95RZ8 B4QBL1 A0A0Q9WCE6 A0A195FKV7 E2A170 A0A0Q9XMD3 A0A0P4W687 K7JCG6 F4WEE6 A0A182F1Y5 A0A1B6L459 A0A0P4W8Y7
PDB
3QWW
E-value=2.66314e-12,
Score=176
Ontologies
GO
Topology
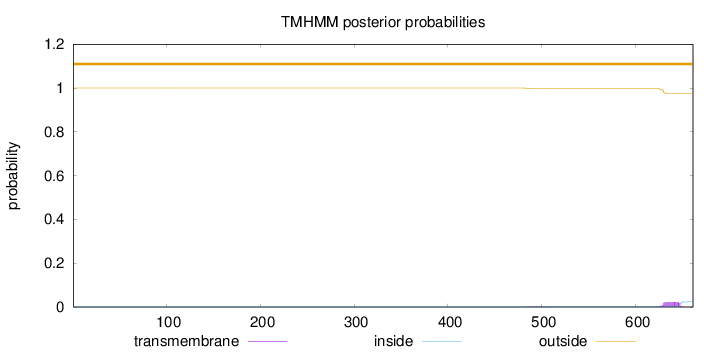
Length:
661
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.45549
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00029
outside
1 - 661
Population Genetic Test Statistics
Pi
349.368956
Theta
213.748268
Tajima's D
1.561969
CLR
0.335456
CSRT
0.80265986700665
Interpretation
Uncertain
