Gene
KWMTBOMO08737
Pre Gene Modal
BGIBMGA007674
Annotation
putative_dynein_light_chain_Tctex-type_[Danaus_plexippus]
Full name
Flavin-containing monooxygenase
Location in the cell
Nuclear Reliability : 2.463
Sequence
CDS
ATGGCCGATGAGAATGTTGTTGAAGAAGAGGCAGAAACACCTGCAGCTACAGAAAGTGAGTCAGCGATAACCCCTACTGAATTGGACAAACCATTGGCTCCACCTAAATATGAAGTGCGCCCTTCTTTGGGCGAAAAATTTCAGTCTCAAAATGTTCGCGAGATCATCGTAACGACAATGCAGGAACAGTTGATGGGTAGAAAGTATAGGTCGGACCAAGCGCCGCGATGGACAGAGGAAATCGCTAACAGAGTTCGACAAAGAGTTCAGGACTTGGATATGAAAAGGTACAAAATTTTGGTACAGTGCATCATGATAGAAATGAAAGGCGCCGGGGTGAAATGCGCCCAGCGCTGCATGTGGGATCCGGAAACGGATGACTATGTCGGCGATTTATTCAGAAATGAAACTATATATTGTTACACTGTTGCGTACGGCATATATATGTATTAA
Protein
MADENVVEEEAETPAATESESAITPTELDKPLAPPKYEVRPSLGEKFQSQNVREIIVTTMQEQLMGRKYRSDQAPRWTEEIANRVRQRVQDLDMKRYKILVQCIMIEMKGAGVKCAQRCMWDPETDDYVGDLFRNETIYCYTVAYGIYMY
Summary
Cofactor
FAD
Similarity
Belongs to the FMO family.
Uniprot
A0A212ERI9
A0A3S2M883
A0A194QMS5
A0A194PQ65
S4Q040
A0A2H1V0D8
+ More
A0A2W1BID5 A0A2A4JA47 A0A1Y1M5H3 D6WD93 A0A067RI58 A0A182NSH1 E0VWR3 A0A084VDD2 A0A2B4RP42 A0A1B6CVW6 A0A182M119 A0A182JTY6 A0A182XFB5 A0A182HYK7 A0A182Y8V5 A0A182SRT5 E9H2V7 A0A0M9A168 A0A182PTB4 A0A182RT44 A0A195FR14 H2Z9D3 A0A2J7RJ30 A0A0P5K1J6 F6THA9 A0A158NAB1 A0A182W3D1 A0A026WL35 A0A2M4AZB6 A0A433UBG2 A0A2M4C2J2 W5J7T7 A0A1Q3FCE9 A0A088A2X6 A0A086Q9G7 Q0C784 A0A151H7P3 S7WFN7 A0A086JII9 A0A125YGW5 A0A086JRA2 A0A2T6IIG9 A0A3R8AMR5 A0A086KPA5 A0A125YGW6 A0A086QIF5 A0A086LPF7 A0A0P5F8A9 A0A2C9KTG2 A0A0G4J5T1 A0A369S3A4 E9IF69 A0A0M0J3B5 A0A2M3ZKF1 A0A423TT54 A0A139XNR7 A0A1W7R905 A0A2G8XQD9 A0A2J8ACT9 A0A154PLI5 A0A195DLW7 Q6IET4 A0A1Y2ED93 D8TMI5 A0A074SX54 A0A195CDP2 A0A150GWM5 K1PA73 A0A0A9Z8T5 K7J1S0 A0A2P6NQZ6 T2M8P7 A0A0L0S4L3 A0A0G4EWI1 A0A195BJC9 V4AYK3 A0A388L0P1 A0A170Y8C5 F4WN78 A0A224XNY7 T1GHC7 A0A0P4VMZ9 A0A1Y2A4V5 A0A1B6E1Y3 A0A397JLG9 A0A2T7NBU2 A0A0V0GB61 A0A1Y3NK27 A0A0L0SUE3 F2U6L1 K4FTH6 A0A1L8GA99 A0A1Y1VHK8
A0A2W1BID5 A0A2A4JA47 A0A1Y1M5H3 D6WD93 A0A067RI58 A0A182NSH1 E0VWR3 A0A084VDD2 A0A2B4RP42 A0A1B6CVW6 A0A182M119 A0A182JTY6 A0A182XFB5 A0A182HYK7 A0A182Y8V5 A0A182SRT5 E9H2V7 A0A0M9A168 A0A182PTB4 A0A182RT44 A0A195FR14 H2Z9D3 A0A2J7RJ30 A0A0P5K1J6 F6THA9 A0A158NAB1 A0A182W3D1 A0A026WL35 A0A2M4AZB6 A0A433UBG2 A0A2M4C2J2 W5J7T7 A0A1Q3FCE9 A0A088A2X6 A0A086Q9G7 Q0C784 A0A151H7P3 S7WFN7 A0A086JII9 A0A125YGW5 A0A086JRA2 A0A2T6IIG9 A0A3R8AMR5 A0A086KPA5 A0A125YGW6 A0A086QIF5 A0A086LPF7 A0A0P5F8A9 A0A2C9KTG2 A0A0G4J5T1 A0A369S3A4 E9IF69 A0A0M0J3B5 A0A2M3ZKF1 A0A423TT54 A0A139XNR7 A0A1W7R905 A0A2G8XQD9 A0A2J8ACT9 A0A154PLI5 A0A195DLW7 Q6IET4 A0A1Y2ED93 D8TMI5 A0A074SX54 A0A195CDP2 A0A150GWM5 K1PA73 A0A0A9Z8T5 K7J1S0 A0A2P6NQZ6 T2M8P7 A0A0L0S4L3 A0A0G4EWI1 A0A195BJC9 V4AYK3 A0A388L0P1 A0A170Y8C5 F4WN78 A0A224XNY7 T1GHC7 A0A0P4VMZ9 A0A1Y2A4V5 A0A1B6E1Y3 A0A397JLG9 A0A2T7NBU2 A0A0V0GB61 A0A1Y3NK27 A0A0L0SUE3 F2U6L1 K4FTH6 A0A1L8GA99 A0A1Y1VHK8
EC Number
1.-.-.-
Pubmed
22118469
26354079
23622113
28756777
28004739
18362917
+ More
19820115 24845553 20566863 24438588 25244985 21292972 12481130 15114417 21347285 24508170 30249741 20920257 23761445 25480939 17510324 26738725 15562597 30042472 21282665 29294063 15020587 17932292 20616280 27102219 22992520 25401762 26823975 20075255 29378020 24065732 23254933 30007417 21719571 27129103 23056606 27762356
19820115 24845553 20566863 24438588 25244985 21292972 12481130 15114417 21347285 24508170 30249741 20920257 23761445 25480939 17510324 26738725 15562597 30042472 21282665 29294063 15020587 17932292 20616280 27102219 22992520 25401762 26823975 20075255 29378020 24065732 23254933 30007417 21719571 27129103 23056606 27762356
EMBL
AGBW02013012
OWR44095.1
RSAL01000014
RVE53245.1
KQ461195
KPJ06649.1
+ More
KQ459596 KPI95457.1 GAIX01000453 JAA92107.1 ODYU01000077 SOQ34226.1 KZ150318 PZC71433.1 NWSH01002176 PCG68965.1 GEZM01040644 JAV80781.1 KQ971321 EEZ99491.2 KK852666 KDR18945.1 DS235824 EEB17819.1 ATLV01011260 KE524659 KFB35976.1 LSMT01000384 PFX18946.1 GEDC01019730 GEDC01001474 JAS17568.1 JAS35824.1 AXCM01000209 APCN01005389 GL732587 EFX73852.1 KQ435768 KOX75250.1 KQ981305 KYN43030.1 NEVH01003009 PNF40830.1 GDIQ01204468 GDIQ01191081 GDIP01117515 GDIP01101990 LRGB01001663 JAK60644.1 JAL86199.1 JAM01725.1 KZS10854.1 EAAA01002404 ADTU01010124 KK107167 QOIP01000010 EZA56381.1 RLU18147.1 GGFK01012799 MBW46120.1 RQTK01000018 RUS91112.1 GGFJ01010364 MBW59505.1 ADMH02001999 ETN60041.1 GFDL01009791 JAV25254.1 AEYJ02000554 KFH09249.1 CH477186 EAT48942.1 AHZP02001997 KYK65328.1 AAQM03000064 EPR63158.1 AEYI02001907 KFG31957.1 KE138840 EPT25286.1 AHZU02001227 KFG34670.1 AFHV02002904 PUA85096.1 AHIV02001323 RQX70409.1 AEYH02001846 KFG46223.1 AAYL02000064 ESS34601.1 AEXC02001521 KFH12387.1 AFYV02002497 KFG58525.1 GDIQ01258701 JAJ93023.1 CDSF01000133 OVEO01000003 CEP02606.1 SPQ94739.1 NOWV01000088 RDD40894.1 GL762758 EFZ20782.1 JWZX01003393 KOO21044.1 GGFM01008204 MBW28955.1 QCYY01001210 ROT79646.1 AGQS02005469 KYF40426.1 GEHC01000025 JAV47620.1 AGQR02003256 PIL97235.1 PGGS01000060 PNH10327.1 KQ434946 KZC12314.1 KQ980734 KYN13890.1 BK004867 DS496130 CM008970 DAA05278.1 EDP02345.1 PNW78911.1 MCOG01000045 ORY69236.1 GL378328 EFJ51127.1 KL544050 KEP62310.1 KQ978009 KYM98178.1 LSYV01000006 KXZ54195.1 JH817363 EKC20657.1 GBHO01002745 GBRD01003958 GDHC01021679 JAG40859.1 JAG61863.1 JAP96949.1 MDYQ01000032 PRP86389.1 HAAD01002189 CDG68421.1 GG745331 KNE57395.1 CDMY01000338 CEM03322.1 KQ976455 KYM85209.1 KB201205 ESO98751.1 BFEA01000231 GBG75802.1 GEMB01003555 JAR99662.1 GL888235 EGI64347.1 GFTR01002231 JAW14195.1 CAQQ02152919 GDKW01003472 JAI53123.1 MCOG01000326 ORY17549.1 GEDC01015003 GEDC01005355 JAS22295.1 JAS31943.1 PQFF01000016 RHZ89205.1 PZQS01000014 PVD18638.1 GECL01001453 JAP04671.1 KZ150777 OUM68129.1 GG745349 KNE66137.1 GL832963 EGD83493.1 JX052857 AFK11085.1 CM004474 OCT80646.1 MCFH01000008 ORX55864.1
KQ459596 KPI95457.1 GAIX01000453 JAA92107.1 ODYU01000077 SOQ34226.1 KZ150318 PZC71433.1 NWSH01002176 PCG68965.1 GEZM01040644 JAV80781.1 KQ971321 EEZ99491.2 KK852666 KDR18945.1 DS235824 EEB17819.1 ATLV01011260 KE524659 KFB35976.1 LSMT01000384 PFX18946.1 GEDC01019730 GEDC01001474 JAS17568.1 JAS35824.1 AXCM01000209 APCN01005389 GL732587 EFX73852.1 KQ435768 KOX75250.1 KQ981305 KYN43030.1 NEVH01003009 PNF40830.1 GDIQ01204468 GDIQ01191081 GDIP01117515 GDIP01101990 LRGB01001663 JAK60644.1 JAL86199.1 JAM01725.1 KZS10854.1 EAAA01002404 ADTU01010124 KK107167 QOIP01000010 EZA56381.1 RLU18147.1 GGFK01012799 MBW46120.1 RQTK01000018 RUS91112.1 GGFJ01010364 MBW59505.1 ADMH02001999 ETN60041.1 GFDL01009791 JAV25254.1 AEYJ02000554 KFH09249.1 CH477186 EAT48942.1 AHZP02001997 KYK65328.1 AAQM03000064 EPR63158.1 AEYI02001907 KFG31957.1 KE138840 EPT25286.1 AHZU02001227 KFG34670.1 AFHV02002904 PUA85096.1 AHIV02001323 RQX70409.1 AEYH02001846 KFG46223.1 AAYL02000064 ESS34601.1 AEXC02001521 KFH12387.1 AFYV02002497 KFG58525.1 GDIQ01258701 JAJ93023.1 CDSF01000133 OVEO01000003 CEP02606.1 SPQ94739.1 NOWV01000088 RDD40894.1 GL762758 EFZ20782.1 JWZX01003393 KOO21044.1 GGFM01008204 MBW28955.1 QCYY01001210 ROT79646.1 AGQS02005469 KYF40426.1 GEHC01000025 JAV47620.1 AGQR02003256 PIL97235.1 PGGS01000060 PNH10327.1 KQ434946 KZC12314.1 KQ980734 KYN13890.1 BK004867 DS496130 CM008970 DAA05278.1 EDP02345.1 PNW78911.1 MCOG01000045 ORY69236.1 GL378328 EFJ51127.1 KL544050 KEP62310.1 KQ978009 KYM98178.1 LSYV01000006 KXZ54195.1 JH817363 EKC20657.1 GBHO01002745 GBRD01003958 GDHC01021679 JAG40859.1 JAG61863.1 JAP96949.1 MDYQ01000032 PRP86389.1 HAAD01002189 CDG68421.1 GG745331 KNE57395.1 CDMY01000338 CEM03322.1 KQ976455 KYM85209.1 KB201205 ESO98751.1 BFEA01000231 GBG75802.1 GEMB01003555 JAR99662.1 GL888235 EGI64347.1 GFTR01002231 JAW14195.1 CAQQ02152919 GDKW01003472 JAI53123.1 MCOG01000326 ORY17549.1 GEDC01015003 GEDC01005355 JAS22295.1 JAS31943.1 PQFF01000016 RHZ89205.1 PZQS01000014 PVD18638.1 GECL01001453 JAP04671.1 KZ150777 OUM68129.1 GG745349 KNE66137.1 GL832963 EGD83493.1 JX052857 AFK11085.1 CM004474 OCT80646.1 MCFH01000008 ORX55864.1
Proteomes
UP000007151
UP000283053
UP000053240
UP000053268
UP000218220
UP000007266
+ More
UP000027135 UP000075884 UP000009046 UP000030765 UP000225706 UP000075883 UP000075881 UP000076407 UP000075840 UP000076408 UP000075901 UP000000305 UP000053105 UP000075885 UP000075900 UP000078541 UP000007875 UP000235965 UP000076858 UP000008144 UP000005205 UP000075920 UP000053097 UP000279307 UP000271974 UP000000673 UP000005203 UP000008820 UP000005641 UP000001529 UP000002226 UP000076420 UP000039324 UP000290189 UP000253843 UP000283509 UP000076502 UP000078492 UP000006906 UP000193920 UP000001058 UP000027470 UP000078542 UP000075714 UP000005408 UP000002358 UP000241769 UP000054350 UP000041254 UP000078540 UP000030746 UP000265515 UP000007755 UP000015102 UP000266861 UP000245119 UP000195826 UP000007799 UP000186698 UP000193719
UP000027135 UP000075884 UP000009046 UP000030765 UP000225706 UP000075883 UP000075881 UP000076407 UP000075840 UP000076408 UP000075901 UP000000305 UP000053105 UP000075885 UP000075900 UP000078541 UP000007875 UP000235965 UP000076858 UP000008144 UP000005205 UP000075920 UP000053097 UP000279307 UP000271974 UP000000673 UP000005203 UP000008820 UP000005641 UP000001529 UP000002226 UP000076420 UP000039324 UP000290189 UP000253843 UP000283509 UP000076502 UP000078492 UP000006906 UP000193920 UP000001058 UP000027470 UP000078542 UP000075714 UP000005408 UP000002358 UP000241769 UP000054350 UP000041254 UP000078540 UP000030746 UP000265515 UP000007755 UP000015102 UP000266861 UP000245119 UP000195826 UP000007799 UP000186698 UP000193719
Interpro
Gene 3D
ProteinModelPortal
A0A212ERI9
A0A3S2M883
A0A194QMS5
A0A194PQ65
S4Q040
A0A2H1V0D8
+ More
A0A2W1BID5 A0A2A4JA47 A0A1Y1M5H3 D6WD93 A0A067RI58 A0A182NSH1 E0VWR3 A0A084VDD2 A0A2B4RP42 A0A1B6CVW6 A0A182M119 A0A182JTY6 A0A182XFB5 A0A182HYK7 A0A182Y8V5 A0A182SRT5 E9H2V7 A0A0M9A168 A0A182PTB4 A0A182RT44 A0A195FR14 H2Z9D3 A0A2J7RJ30 A0A0P5K1J6 F6THA9 A0A158NAB1 A0A182W3D1 A0A026WL35 A0A2M4AZB6 A0A433UBG2 A0A2M4C2J2 W5J7T7 A0A1Q3FCE9 A0A088A2X6 A0A086Q9G7 Q0C784 A0A151H7P3 S7WFN7 A0A086JII9 A0A125YGW5 A0A086JRA2 A0A2T6IIG9 A0A3R8AMR5 A0A086KPA5 A0A125YGW6 A0A086QIF5 A0A086LPF7 A0A0P5F8A9 A0A2C9KTG2 A0A0G4J5T1 A0A369S3A4 E9IF69 A0A0M0J3B5 A0A2M3ZKF1 A0A423TT54 A0A139XNR7 A0A1W7R905 A0A2G8XQD9 A0A2J8ACT9 A0A154PLI5 A0A195DLW7 Q6IET4 A0A1Y2ED93 D8TMI5 A0A074SX54 A0A195CDP2 A0A150GWM5 K1PA73 A0A0A9Z8T5 K7J1S0 A0A2P6NQZ6 T2M8P7 A0A0L0S4L3 A0A0G4EWI1 A0A195BJC9 V4AYK3 A0A388L0P1 A0A170Y8C5 F4WN78 A0A224XNY7 T1GHC7 A0A0P4VMZ9 A0A1Y2A4V5 A0A1B6E1Y3 A0A397JLG9 A0A2T7NBU2 A0A0V0GB61 A0A1Y3NK27 A0A0L0SUE3 F2U6L1 K4FTH6 A0A1L8GA99 A0A1Y1VHK8
A0A2W1BID5 A0A2A4JA47 A0A1Y1M5H3 D6WD93 A0A067RI58 A0A182NSH1 E0VWR3 A0A084VDD2 A0A2B4RP42 A0A1B6CVW6 A0A182M119 A0A182JTY6 A0A182XFB5 A0A182HYK7 A0A182Y8V5 A0A182SRT5 E9H2V7 A0A0M9A168 A0A182PTB4 A0A182RT44 A0A195FR14 H2Z9D3 A0A2J7RJ30 A0A0P5K1J6 F6THA9 A0A158NAB1 A0A182W3D1 A0A026WL35 A0A2M4AZB6 A0A433UBG2 A0A2M4C2J2 W5J7T7 A0A1Q3FCE9 A0A088A2X6 A0A086Q9G7 Q0C784 A0A151H7P3 S7WFN7 A0A086JII9 A0A125YGW5 A0A086JRA2 A0A2T6IIG9 A0A3R8AMR5 A0A086KPA5 A0A125YGW6 A0A086QIF5 A0A086LPF7 A0A0P5F8A9 A0A2C9KTG2 A0A0G4J5T1 A0A369S3A4 E9IF69 A0A0M0J3B5 A0A2M3ZKF1 A0A423TT54 A0A139XNR7 A0A1W7R905 A0A2G8XQD9 A0A2J8ACT9 A0A154PLI5 A0A195DLW7 Q6IET4 A0A1Y2ED93 D8TMI5 A0A074SX54 A0A195CDP2 A0A150GWM5 K1PA73 A0A0A9Z8T5 K7J1S0 A0A2P6NQZ6 T2M8P7 A0A0L0S4L3 A0A0G4EWI1 A0A195BJC9 V4AYK3 A0A388L0P1 A0A170Y8C5 F4WN78 A0A224XNY7 T1GHC7 A0A0P4VMZ9 A0A1Y2A4V5 A0A1B6E1Y3 A0A397JLG9 A0A2T7NBU2 A0A0V0GB61 A0A1Y3NK27 A0A0L0SUE3 F2U6L1 K4FTH6 A0A1L8GA99 A0A1Y1VHK8
PDB
1XDX
E-value=1.01432e-08,
Score=137
Ontologies
GO
Topology
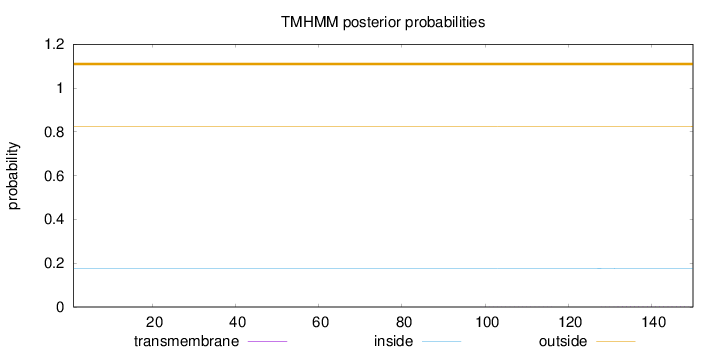
Length:
150
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03057
Exp number, first 60 AAs:
0
Total prob of N-in:
0.17545
outside
1 - 150
Population Genetic Test Statistics
Pi
169.833353
Theta
191.135212
Tajima's D
1.238487
CLR
0.723496
CSRT
0.714164291785411
Interpretation
Uncertain
