Gene
KWMTBOMO08734
Pre Gene Modal
BGIBMGA007909
Annotation
PREDICTED:_rab_GTPase-activating_protein_1-like_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.934
Sequence
CDS
ATGGAGTTCAATGAATATCAGACAAGACAAGTCTCGCATTCAGTAGATGTTTATTCTACCGTTCGTTCACGGTCGCAAAGTTCCTCACCTGTGCTTTGTACGAATGTGTCTTGCGATTTGGAGGACATTTTGAGCGACGCCATCGCAGAAACATTACAAAGTTCTCTCCGTGTAAGTCACGGTCAGTTAGTGAGCCAGGAACTTTCTATTCGACCAGAAGCCAATATTGAAGCCAACGTTCCAGAAGATCTTGATTTATTTCAAGAAATATTTGAAGTTGATAGTGAGTCTGAACCAAGAATGGGAGATTCTAATGGAGCTCCAGTGCAGAACAATGTATTGAAAGAGCAGGATTCGTGTCGTCTGGTCGGTGGCTCAAAGTTCTATGAAATGGTCGATGGCTCGCCATCTGCCGATACGTCCACACCGTCACCAACAGAGAAGCATGATCATATGAAGCCGATAGAGAGCGACGATGAAGAATGCACAGTGTTCAGCGGCGTCAGTTACCTGGGAGCCCAGAACATTTCGGATCCGAAGTGCGAAAGCGACATCCAGCGGATTATGAAGGAACTAAGTTCGATGCCTGAGAGCAGGGACGGGATCGCAGTGTCGATATCGATACCTGTGTGTTCGCAGGGATTGGTTGTGCTGTACCAGGCGGATTCTAATAGCGTGATGGCGCGCTACGCCGTCAATCGCATTTCGTTCTACGCTCGCGGCGCGGTCGGTTCGCCGGTCGCTTCGTGCTTCGCGTTCACCTGGTCCCACGGCGAGACCAAGGAATCCGCAGTGTACAGGTGCCACGTGTTCCGCTGTCACATCGCTGAAGCTGTCACACAAGTGTCAAGTTGCTTCGCCAAGGCTTTCGAGCGCGTTCCTAGATCTATGACGTCATCAATAGCTGGTGAGCTGAGCGCGGCGCCCTCTATGACCGGTTCGCTTATTGAAGGTCTCGGACCCGCCGCTCCACCGATGCAGCTCATGCACGTATTGGAATGCACCGTTGATATCAAGGAAGCGGATGCTAAGGGAACTTTTAGCCATGTTCCGAAGGAGCGTCTCGGTTTCAAGCTGCGTGGTGGCATCGACAAGCAAGTGACCATCAATGTCCAACAGGTCACTAATAATGTTCAGACTGAAGTAGACGAGGCACAGAGTGGCTACACCGGCGGCTTGTACATCGAAAGATGCTTTGGGATACTCCTGGCTCCGGGGCGCAGCGTTAAACATTCAGACATGAGACTGCTAGAAATGGTGAGCGGTTCATCGGGAGGTTCGGGTTGTTCGGTGTGCGCGCTGTGGAACGCGGGGGAGTCCGCACTCGCGGCCTTCAACGTCGCCAGCGGGGACGGGGCTCCGGCGTACATGTCTCTGGCGCTCGACCTCGTCATACGGGGCATCCCTGACCCCCTCAGGTTGATCATAGAAACTCCGGTGAAAGTGTTTCCGCCAACAGAAAGGTTTTGGTATTATTCGAAAAGACCTCTCGTTCAACAGTTTTACATTAATCTTAAAGAGTCCGTGGATCCTCTGGGTCAGCTCCAGTACGAGGTGGCGAGCGTGGAGACCGAGGGGGAGCTGGACAAGTCCCGTTTGAACTTGCCGCTGTCGCTGTCCAGCCTGCTGCCGTCGCCGCTGTCCAGCGTGCCCGCCCCGCCCTCGCCCTCCGAACCGGACTCTGATGGCGATGAGCCGCTTCTGAGTGGAACTGGAGAAGTTTCTAAAGATTGCGGCGAAGATGTGCTCGTAAACTGGGCCCAGGTGCTAAGAAGTTGGACTGGCCCGAGACCCAGAGCATTGAACCACTTGGTGCGGGTCGGTGTGCCCGAAGCCCTCCGCGGGGAGGTGTGGCTCCGGCTGGCCGGCGTCGACCAGAACGATAAACTAATGGAGACCTACAGAACGTTAATTAGTAAGGAGTGTCCGTTCGAGGCGGTAATCCAGCGCGACATCGCGCGCACATTCCCGGCGCACGACTTCTTCCGCGAGGCGGGCGGGCTCGGGCAGGACTCGCTGCTGCGGATGGCGCGCGCCTACGCCGTGTACGACCACGAGGTCGGCTACTGCCAAGGACTCAGCTTCCTGGCCGCCACGCTGCTGCTGCACATGCCTGAAGAGCAAGCTTTCTGCCTTCTTGTCCGGCTAATGTATGGCTATGGACTAAGAGAGCTATACAAGGACGGATTTGAAGCTCTCTATATGAGACTGCATCAACTAGATAGGCTGATCGAGGAACAATTGACGGACTTACGTGCGCATTTCCAAGAACTAGGTGTCGAGCCGCACATGTTTGCATCGCAATGGTTCCTAACGGTGTTCACTGCACGCTTCCCTCTACCATTGGTGTACCACATTCTAGACGTGTTTCTGCTGCAAGGTATCGACACGTTATTCCAAGTTGCACTGGCGTTGCTCTCTAGGTCACGGAAGGACTTACTGACACACGATTTCGAAGGTGTCCTGAAATATTTCAGAGTAACGCTGCCTAAGAAATGTCGCGCTGAGGAGACATCGAGACAGATCATCAAACTTGCGTGCAGCATCAAAGTGAAACGACTCAACAAATACCAGTTGGAATATGAGAAGTTTATCAAAGAATCTGCTGAAAAAGAGAAAATAAATGTAGAAATGGAACGATTGAAAGTAATCAACAATCAACTGCAACTAGAGAAAGAAACGCTCGAAACCCAATTGAACGAGGCCCGCGCTAATTACGAAGAAGCCGAGAAGGCGACGGCGCACTACGCGTCCGTTGCCAGGGACTACAGAGAGATCTGCGCCAGGCTCGACAGGCAGCTGCACGTCCTGCAGGACACCGTAAAAGATTGTGTAACTTGCAGCATGAAATTGACACCGAAGGATAATAAAGAAGCAGAAAAAGGCGACGGCAATCTCGAAGGCACAACGGATACTGAGACTAGATTAAGACAAAAAATATGCGACTTGGAACTCGAACTAGCTCAAGTGAAACTGGCGCACGTACAGGCGCAGTGCAGTAATCAGGAACTCAGTCACCAGCTTAGTGCAGCTCTGAACGAGGTGCAAGCCGCGATGAACAACCAAAAGCAAACCGTCGCACCTTGGCTAGTTCGAACACTTGGCTCCATAATGGACGCGGCACAGAATAAATCAACATTCCAGACGTACATACCGAATTTTAATCAAGAATCTTCAACGCCCATTCATCGACAAGGCTCGTTTTCTAGTCAAAGCCACAATCCAGTGAACCTCGACAGACGAGTCTCTGAGCCGTACAGTCCCGATGTTGTCCGAAGGAAGAAGGACCTCAACGCCAAGAGACATTCGATGCTCGTCGAGTCGAACATGACCAAAGAGGCCAAAGAAATGAATCGGCGAAGTCATGACGTCACGATAGTGAAGAACATCTCGATGGGTTGA
Protein
MEFNEYQTRQVSHSVDVYSTVRSRSQSSSPVLCTNVSCDLEDILSDAIAETLQSSLRVSHGQLVSQELSIRPEANIEANVPEDLDLFQEIFEVDSESEPRMGDSNGAPVQNNVLKEQDSCRLVGGSKFYEMVDGSPSADTSTPSPTEKHDHMKPIESDDEECTVFSGVSYLGAQNISDPKCESDIQRIMKELSSMPESRDGIAVSISIPVCSQGLVVLYQADSNSVMARYAVNRISFYARGAVGSPVASCFAFTWSHGETKESAVYRCHVFRCHIAEAVTQVSSCFAKAFERVPRSMTSSIAGELSAAPSMTGSLIEGLGPAAPPMQLMHVLECTVDIKEADAKGTFSHVPKERLGFKLRGGIDKQVTINVQQVTNNVQTEVDEAQSGYTGGLYIERCFGILLAPGRSVKHSDMRLLEMVSGSSGGSGCSVCALWNAGESALAAFNVASGDGAPAYMSLALDLVIRGIPDPLRLIIETPVKVFPPTERFWYYSKRPLVQQFYINLKESVDPLGQLQYEVASVETEGELDKSRLNLPLSLSSLLPSPLSSVPAPPSPSEPDSDGDEPLLSGTGEVSKDCGEDVLVNWAQVLRSWTGPRPRALNHLVRVGVPEALRGEVWLRLAGVDQNDKLMETYRTLISKECPFEAVIQRDIARTFPAHDFFREAGGLGQDSLLRMARAYAVYDHEVGYCQGLSFLAATLLLHMPEEQAFCLLVRLMYGYGLRELYKDGFEALYMRLHQLDRLIEEQLTDLRAHFQELGVEPHMFASQWFLTVFTARFPLPLVYHILDVFLLQGIDTLFQVALALLSRSRKDLLTHDFEGVLKYFRVTLPKKCRAEETSRQIIKLACSIKVKRLNKYQLEYEKFIKESAEKEKINVEMERLKVINNQLQLEKETLETQLNEARANYEEAEKATAHYASVARDYREICARLDRQLHVLQDTVKDCVTCSMKLTPKDNKEAEKGDGNLEGTTDTETRLRQKICDLELELAQVKLAHVQAQCSNQELSHQLSAALNEVQAAMNNQKQTVAPWLVRTLGSIMDAAQNKSTFQTYIPNFNQESSTPIHRQGSFSSQSHNPVNLDRRVSEPYSPDVVRRKKDLNAKRHSMLVESNMTKEAKEMNRRSHDVTIVKNISMG
Summary
Uniprot
A0A3S2P5Z9
A0A212ERJ1
A0A194QNU4
A0A194PR68
H9JEG2
A0A2A4JAU4
+ More
A0A2W1BDP8 A0A2J7Q0H9 A0A2J7Q0I6 D6X3W7 A0A067RJX1 A0A1Y1JTN6 A0A154PJM8 A0A1Y1JTN7 A0A224XAP6 A0A069DWY1 A0A026WMF8 A0A023F259 A0A195BEC7 A0A195DI05 A0A195EQV6 A0A088ACG3 A0A1B6CKT3 A0A1B6D067 E1ZXC8 E2B8J0 K7IRD5 T1HQL1 A0A232F0X2 A0A146M2Z8 A0A0A9XPR8 A0A0A9XT17 A0A146M341 E0VTD1 A0A1B6HUV9 A0A158NQS4 A0A0A9XQR2 A0A146LJE7 A0A0A9XML6 A0A0K8SRU6 A0A1B6FZ96 A0A1L8DVR7 A0A0K8TSK8 A0A0K8U1S3 A0A034VF56 J9JYT1 A0A0K8UB34 A0A2S2QE07 A0A0A1XRV4 A0A1B0AYA2 B4KYD0 A0A1A9XWI9 A0A0Q9XNJ0 A0A0A1WP96 A0A0P8ZU93 B3M5J8 T1PE13 Q8MT33 Q9VSI2 A0A1W4US74 A0A084WS41 A0A1A9WR95 A0A1Q3F2H4 A0A1A9Z3C5 Q17ND9 Q17ND8 A0A1A9VYM3 A0A182RU23 B0W6W9 F4WBX7 A0A0L8GQZ5 A0A0L8GR89 A0A0L8GRE5 A0A0L8GS12 A0A0L8GQY9 A0A0L8GS16 A0A2L2YA69 A0A1D2NJA9 A0A3Q1LIH2 A0A1L8F6C7 A0A3Q7QQ65 A0A3Q7TUY1
A0A2W1BDP8 A0A2J7Q0H9 A0A2J7Q0I6 D6X3W7 A0A067RJX1 A0A1Y1JTN6 A0A154PJM8 A0A1Y1JTN7 A0A224XAP6 A0A069DWY1 A0A026WMF8 A0A023F259 A0A195BEC7 A0A195DI05 A0A195EQV6 A0A088ACG3 A0A1B6CKT3 A0A1B6D067 E1ZXC8 E2B8J0 K7IRD5 T1HQL1 A0A232F0X2 A0A146M2Z8 A0A0A9XPR8 A0A0A9XT17 A0A146M341 E0VTD1 A0A1B6HUV9 A0A158NQS4 A0A0A9XQR2 A0A146LJE7 A0A0A9XML6 A0A0K8SRU6 A0A1B6FZ96 A0A1L8DVR7 A0A0K8TSK8 A0A0K8U1S3 A0A034VF56 J9JYT1 A0A0K8UB34 A0A2S2QE07 A0A0A1XRV4 A0A1B0AYA2 B4KYD0 A0A1A9XWI9 A0A0Q9XNJ0 A0A0A1WP96 A0A0P8ZU93 B3M5J8 T1PE13 Q8MT33 Q9VSI2 A0A1W4US74 A0A084WS41 A0A1A9WR95 A0A1Q3F2H4 A0A1A9Z3C5 Q17ND9 Q17ND8 A0A1A9VYM3 A0A182RU23 B0W6W9 F4WBX7 A0A0L8GQZ5 A0A0L8GR89 A0A0L8GRE5 A0A0L8GS12 A0A0L8GQY9 A0A0L8GS16 A0A2L2YA69 A0A1D2NJA9 A0A3Q1LIH2 A0A1L8F6C7 A0A3Q7QQ65 A0A3Q7TUY1
Pubmed
22118469
26354079
19121390
28756777
18362917
19820115
+ More
24845553 28004739 26334808 24508170 25474469 20798317 20075255 28648823 26823975 25401762 20566863 21347285 26369729 25348373 25830018 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 24438588 17510324 21719571 26561354 27289101 19393038 27762356
24845553 28004739 26334808 24508170 25474469 20798317 20075255 28648823 26823975 25401762 20566863 21347285 26369729 25348373 25830018 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 24438588 17510324 21719571 26561354 27289101 19393038 27762356
EMBL
RSAL01000014
RVE53242.1
AGBW02013012
OWR44097.1
KQ461195
KPJ06645.1
+ More
KQ459596 KPI95453.1 BABH01003045 BABH01003046 BABH01003047 BABH01003048 NWSH01002328 PCG68513.1 KZ150318 PZC71437.1 NEVH01019967 PNF22092.1 PNF22091.1 KQ971379 EEZ97478.2 KK852463 KDR23318.1 GEZM01103174 JAV51488.1 KQ434923 KZC11488.1 GEZM01103178 JAV51481.1 GFTR01008422 JAW08004.1 GBGD01000306 JAC88583.1 KK107154 EZA56851.1 GBBI01003045 JAC15667.1 KQ976511 KYM82564.1 KQ980824 KYN12528.1 KQ982021 KYN30593.1 GEDC01029621 GEDC01023258 GEDC01023060 GEDC01016398 JAS07677.1 JAS14040.1 JAS14238.1 JAS20900.1 GEDC01018189 GEDC01014524 JAS19109.1 JAS22774.1 GL435030 EFN74218.1 GL446337 EFN87976.1 ACPB03008564 NNAY01001402 OXU24097.1 GDHC01004478 JAQ14151.1 GBHO01021635 JAG21969.1 GBHO01021636 GDHC01013778 JAG21968.1 JAQ04851.1 GDHC01004446 JAQ14183.1 DS235762 EEB16637.1 GECU01029233 GECU01010714 JAS78473.1 JAS96992.1 ADTU01023428 ADTU01023429 ADTU01023430 GBHO01021633 JAG21971.1 GDHC01012069 JAQ06560.1 GBHO01021637 JAG21967.1 GBRD01009801 JAG56023.1 GECZ01014254 JAS55515.1 GFDF01003564 JAV10520.1 GDAI01000673 JAI16930.1 GDHF01032059 GDHF01013576 GDHF01009422 JAI20255.1 JAI38738.1 JAI42892.1 GAKP01018251 GAKP01018250 GAKP01018249 JAC40702.1 ABLF02035135 GDHF01028465 JAI23849.1 GGMS01006718 MBY75921.1 GBXI01003119 GBXI01000657 JAD11173.1 JAD13635.1 JXJN01005582 CH933809 EDW17709.1 KRG05704.1 GBXI01013610 JAD00682.1 CH902618 KPU78106.1 EDV39608.1 KA646997 AFP61626.1 AY118408 AAM48437.1 AE014296 AAF50437.2 ATLV01026323 KE525409 KFB53035.1 GFDL01013289 JAV21756.1 CH477200 EAT48196.1 EAT48197.1 DS231850 EDS37115.1 GL888066 EGI68405.1 KQ420776 KOF79408.1 KOF79407.1 KOF79409.1 KOF79405.1 KOF79403.1 KOF79410.1 IAAA01011057 IAAA01011058 LAA04250.1 LJIJ01000024 ODN05350.1 CM004480 OCT67136.1
KQ459596 KPI95453.1 BABH01003045 BABH01003046 BABH01003047 BABH01003048 NWSH01002328 PCG68513.1 KZ150318 PZC71437.1 NEVH01019967 PNF22092.1 PNF22091.1 KQ971379 EEZ97478.2 KK852463 KDR23318.1 GEZM01103174 JAV51488.1 KQ434923 KZC11488.1 GEZM01103178 JAV51481.1 GFTR01008422 JAW08004.1 GBGD01000306 JAC88583.1 KK107154 EZA56851.1 GBBI01003045 JAC15667.1 KQ976511 KYM82564.1 KQ980824 KYN12528.1 KQ982021 KYN30593.1 GEDC01029621 GEDC01023258 GEDC01023060 GEDC01016398 JAS07677.1 JAS14040.1 JAS14238.1 JAS20900.1 GEDC01018189 GEDC01014524 JAS19109.1 JAS22774.1 GL435030 EFN74218.1 GL446337 EFN87976.1 ACPB03008564 NNAY01001402 OXU24097.1 GDHC01004478 JAQ14151.1 GBHO01021635 JAG21969.1 GBHO01021636 GDHC01013778 JAG21968.1 JAQ04851.1 GDHC01004446 JAQ14183.1 DS235762 EEB16637.1 GECU01029233 GECU01010714 JAS78473.1 JAS96992.1 ADTU01023428 ADTU01023429 ADTU01023430 GBHO01021633 JAG21971.1 GDHC01012069 JAQ06560.1 GBHO01021637 JAG21967.1 GBRD01009801 JAG56023.1 GECZ01014254 JAS55515.1 GFDF01003564 JAV10520.1 GDAI01000673 JAI16930.1 GDHF01032059 GDHF01013576 GDHF01009422 JAI20255.1 JAI38738.1 JAI42892.1 GAKP01018251 GAKP01018250 GAKP01018249 JAC40702.1 ABLF02035135 GDHF01028465 JAI23849.1 GGMS01006718 MBY75921.1 GBXI01003119 GBXI01000657 JAD11173.1 JAD13635.1 JXJN01005582 CH933809 EDW17709.1 KRG05704.1 GBXI01013610 JAD00682.1 CH902618 KPU78106.1 EDV39608.1 KA646997 AFP61626.1 AY118408 AAM48437.1 AE014296 AAF50437.2 ATLV01026323 KE525409 KFB53035.1 GFDL01013289 JAV21756.1 CH477200 EAT48196.1 EAT48197.1 DS231850 EDS37115.1 GL888066 EGI68405.1 KQ420776 KOF79408.1 KOF79407.1 KOF79409.1 KOF79405.1 KOF79403.1 KOF79410.1 IAAA01011057 IAAA01011058 LAA04250.1 LJIJ01000024 ODN05350.1 CM004480 OCT67136.1
Proteomes
UP000283053
UP000007151
UP000053240
UP000053268
UP000005204
UP000218220
+ More
UP000235965 UP000007266 UP000027135 UP000076502 UP000053097 UP000078540 UP000078492 UP000078541 UP000005203 UP000000311 UP000008237 UP000002358 UP000015103 UP000215335 UP000009046 UP000005205 UP000007819 UP000092460 UP000009192 UP000092443 UP000007801 UP000000803 UP000192221 UP000030765 UP000091820 UP000092445 UP000008820 UP000078200 UP000075900 UP000002320 UP000007755 UP000053454 UP000094527 UP000009136 UP000186698 UP000286641 UP000286640
UP000235965 UP000007266 UP000027135 UP000076502 UP000053097 UP000078540 UP000078492 UP000078541 UP000005203 UP000000311 UP000008237 UP000002358 UP000015103 UP000215335 UP000009046 UP000005205 UP000007819 UP000092460 UP000009192 UP000092443 UP000007801 UP000000803 UP000192221 UP000030765 UP000091820 UP000092445 UP000008820 UP000078200 UP000075900 UP000002320 UP000007755 UP000053454 UP000094527 UP000009136 UP000186698 UP000286641 UP000286640
Interpro
SUPFAM
SSF47923
SSF47923
Gene 3D
ProteinModelPortal
A0A3S2P5Z9
A0A212ERJ1
A0A194QNU4
A0A194PR68
H9JEG2
A0A2A4JAU4
+ More
A0A2W1BDP8 A0A2J7Q0H9 A0A2J7Q0I6 D6X3W7 A0A067RJX1 A0A1Y1JTN6 A0A154PJM8 A0A1Y1JTN7 A0A224XAP6 A0A069DWY1 A0A026WMF8 A0A023F259 A0A195BEC7 A0A195DI05 A0A195EQV6 A0A088ACG3 A0A1B6CKT3 A0A1B6D067 E1ZXC8 E2B8J0 K7IRD5 T1HQL1 A0A232F0X2 A0A146M2Z8 A0A0A9XPR8 A0A0A9XT17 A0A146M341 E0VTD1 A0A1B6HUV9 A0A158NQS4 A0A0A9XQR2 A0A146LJE7 A0A0A9XML6 A0A0K8SRU6 A0A1B6FZ96 A0A1L8DVR7 A0A0K8TSK8 A0A0K8U1S3 A0A034VF56 J9JYT1 A0A0K8UB34 A0A2S2QE07 A0A0A1XRV4 A0A1B0AYA2 B4KYD0 A0A1A9XWI9 A0A0Q9XNJ0 A0A0A1WP96 A0A0P8ZU93 B3M5J8 T1PE13 Q8MT33 Q9VSI2 A0A1W4US74 A0A084WS41 A0A1A9WR95 A0A1Q3F2H4 A0A1A9Z3C5 Q17ND9 Q17ND8 A0A1A9VYM3 A0A182RU23 B0W6W9 F4WBX7 A0A0L8GQZ5 A0A0L8GR89 A0A0L8GRE5 A0A0L8GS12 A0A0L8GQY9 A0A0L8GS16 A0A2L2YA69 A0A1D2NJA9 A0A3Q1LIH2 A0A1L8F6C7 A0A3Q7QQ65 A0A3Q7TUY1
A0A2W1BDP8 A0A2J7Q0H9 A0A2J7Q0I6 D6X3W7 A0A067RJX1 A0A1Y1JTN6 A0A154PJM8 A0A1Y1JTN7 A0A224XAP6 A0A069DWY1 A0A026WMF8 A0A023F259 A0A195BEC7 A0A195DI05 A0A195EQV6 A0A088ACG3 A0A1B6CKT3 A0A1B6D067 E1ZXC8 E2B8J0 K7IRD5 T1HQL1 A0A232F0X2 A0A146M2Z8 A0A0A9XPR8 A0A0A9XT17 A0A146M341 E0VTD1 A0A1B6HUV9 A0A158NQS4 A0A0A9XQR2 A0A146LJE7 A0A0A9XML6 A0A0K8SRU6 A0A1B6FZ96 A0A1L8DVR7 A0A0K8TSK8 A0A0K8U1S3 A0A034VF56 J9JYT1 A0A0K8UB34 A0A2S2QE07 A0A0A1XRV4 A0A1B0AYA2 B4KYD0 A0A1A9XWI9 A0A0Q9XNJ0 A0A0A1WP96 A0A0P8ZU93 B3M5J8 T1PE13 Q8MT33 Q9VSI2 A0A1W4US74 A0A084WS41 A0A1A9WR95 A0A1Q3F2H4 A0A1A9Z3C5 Q17ND9 Q17ND8 A0A1A9VYM3 A0A182RU23 B0W6W9 F4WBX7 A0A0L8GQZ5 A0A0L8GR89 A0A0L8GRE5 A0A0L8GS12 A0A0L8GQY9 A0A0L8GS16 A0A2L2YA69 A0A1D2NJA9 A0A3Q1LIH2 A0A1L8F6C7 A0A3Q7QQ65 A0A3Q7TUY1
PDB
3HZJ
E-value=1.63734e-96,
Score=905
Ontologies
KEGG
GO
PANTHER
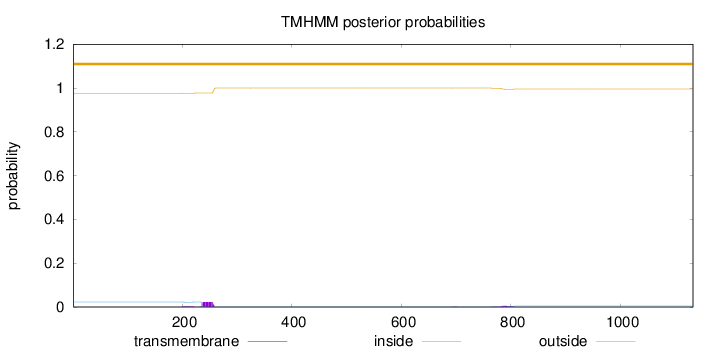
Topology
Length:
1133
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.657709999999997
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02340
outside
1 - 1133
Population Genetic Test Statistics
Pi
272.189507
Theta
168.865452
Tajima's D
1.469697
CLR
0.366683
CSRT
0.776561171941403
Interpretation
Uncertain
