Gene
KWMTBOMO08732
Annotation
PREDICTED:_uncharacterized_protein_LOC101737183_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.218 PlasmaMembrane Reliability : 1.24
Sequence
CDS
ATGACTTCGCGCGGGAAAGTCATTCAAAATGTCCCTAACTATTGTCGGATCGCTCCCGCTATGGACGCGCCGCCGGACTACGACGGCGAAGTCCCTGCTGAGGTGCCCCATATAACGGGGGCGGAGCTCCGTGTGGCCGTTCGCAAAATGTGCGCGAAGGACACTGCACCCGGCCCGGATGGCGTCCATGGCCGGGTTTGGGCCTTGGCTCTTGGTGCCCTAGGGGACCGACTCTTGAGACTTTACAACTCCTGCCTCGAGTCGGGACGGTTTCCTTCATCGTGGAGGACGGTTAGACTCGTGCTGTTGAGAAAGGAGGGGCGCCCGGCGGATACTGCCGCCGGGTACCGTCCCATCGTGTTGCTGGATGAGGTGGGCAAGCTGCTGGAACGCATTCTGGCAGCCCGCATCATCCAGCATCTGGTCGGGGTGGGACCCGATCTGTCGGCGGAGCAGTATGGCTTCCGAGAGGGCCGCTCAACGGTAGACGCGATCTTTCGCGTGCGGGCCCTCTCGGAGGAGGCCGTCTCCAGGGGTGGGGTGGCTTTGGCGGTGTCGCTCGATATCGCCAATGCGTTTAACACCCTCCCCTGGTCCGTGATAGGGGGGGCGCTGGAACGACATAGAGTGCCCCCCTACCTTCGCCGGCTGGTGGGTTCCTACTTAGAGGACAGATCGGTCACGTGTACCGGACATGGTGGGATCCTGCACCGGTTCCCGGTCGTGCGCGGTGTTCCACAGGGGTCGGTGCTCGGCCCCCTTTTGTGGAATATCGGGTATGACTGGGTGCTGAGAGGTGCCCTCCTCCCGGGCCTGAGCGTAATATGTTACGCGGACGACACGTTGGTCGTGGCCCGGGGGGGAGTTTTGCTGAGTCTGCCCGTCTTGCTACGGCTGGGGCCCCGGAGAGTGCCACCTGTCGATGCCCATATCGTGGTTGGAGGCGTCCGTATCGGGGTCGGGGTGCAGTTGAAGTACCTCGGCCTCATTCTGGACAGTCGTTGGACCTTCCGTGCTCACTTTCAGAACCTGGTCCCTCGTTTGTTGGGGGTGGCCGGCGCGCTAAGCCGGCTTCTGCCCAACGTCGGGGGGCCTGACCAGGTGACGCGCCGTCTCTATACGGGGGTGGTGCGATCAATGGCCCTATACGGGGCGCCCGTGTGGGGCCAGTCCCTGGCCGTGGGGGTGGCGAAGCTGCTGCAACGGCCGCAACGCACCATCGCGGTCAGGGTCATCCGTGGTTATCGCACCATCTCCTTTGAGGCGGCGTGTGTACTGGCTGGGACGCCGCCTTGGGTTCTGGAAGCGGAGGCGCTCGCCGCTGACTATAGGTGGCGGGCTGAACTTCGTGCCCGGGGCGTGGCGCGTCCCAGCCCCAGTGTGGTCAGAGCACGGAGGGCCCAATCTCGGCGGTCCGTGCTGGAGTCATGGTCCAGACGGCTGGCTGATCCTTCGGCTGGTCGTAGGACCGTCGAGGCGATTCGCCCGGTCCTTGTGGATTGGGTGAATCGTGACAGAGGACGCCTCACTTTCCGGCTTACGCAGGTGCTCACTGGGCATGGTTGTTTCGGTGAGTTCCTGCACCGGATCACAGCCGAGCCGACGGCAGAGTGCCACCATTGTGGTTGCGACTTGGACACGGCAGAGCATACGCTCGTCGCCTGCCCCGCATGGGAGGGGTGGCGCCGTGTCCTCGTCGCAAAAATAGGAACCGACTTGTCGTTGCCGAGTGTTGTGGCATCGATGCTCGGCGACGACGAGTCGTGGAAGGCGATGCTCGACTTCTGCGAGTGCACCATCTCGCAGAAGGAGGCGGCGGGGCGCGTGAGAGACGCACAATCCCGCCGCCGTCGAGCGGGGGCCAGGGAGGCGGATCTCGCCCAAGCCCTGGCCCTCTAA
Protein
MTSRGKVIQNVPNYCRIAPAMDAPPDYDGEVPAEVPHITGAELRVAVRKMCAKDTAPGPDGVHGRVWALALGALGDRLLRLYNSCLESGRFPSSWRTVRLVLLRKEGRPADTAAGYRPIVLLDEVGKLLERILAARIIQHLVGVGPDLSAEQYGFREGRSTVDAIFRVRALSEEAVSRGGVALAVSLDIANAFNTLPWSVIGGALERHRVPPYLRRLVGSYLEDRSVTCTGHGGILHRFPVVRGVPQGSVLGPLLWNIGYDWVLRGALLPGLSVICYADDTLVVARGGVLLSLPVLLRLGPRRVPPVDAHIVVGGVRIGVGVQLKYLGLILDSRWTFRAHFQNLVPRLLGVAGALSRLLPNVGGPDQVTRRLYTGVVRSMALYGAPVWGQSLAVGVAKLLQRPQRTIAVRVIRGYRTISFEAACVLAGTPPWVLEAEALAADYRWRAELRARGVARPSPSVVRARRAQSRRSVLESWSRRLADPSAGRRTVEAIRPVLVDWVNRDRGRLTFRLTQVLTGHGCFGEFLHRITAEPTAECHHCGCDLDTAEHTLVACPAWEGWRRVLVAKIGTDLSLPSVVASMLGDDESWKAMLDFCECTISQKEAAGRVRDAQSRRRRAGAREADLAQALAL
Summary
Uniprot
Q868Q4
Q8MY27
Q8MY33
Q8MY31
Q8MY35
A0A2W1C3J5
+ More
A0A0J7N1D9 A0A0J7KS67 A0A0J7N7S8 A0A0J7KLT9 A0A0J7KIY5 A0A2S2N8I2 A0A2S2PBP5 A0A0J7KYD2 X1WJY4 A0A2S2PEZ7 A0A0J7JZ13 A0A0J7NEG9 A0A0J7NAV8 A0A2S2NJ82 A0A2H8TIE5 A0A0J7KL06 A0A0J7KMG5 J9JWJ3 A0A0J7MY17 A0A0J7NMN5 J9LVU7 A0A2S2P488 X1WT13 J9KNS1 A0A2H8TR36 X1WMW4 A0A2S2NQQ6 X1WXT5 J9KNG8 A0A2S2PEU2 J9KNN8 A0A2S2PRL8 T1DCM0 A0A142LX45 X1WSG8 A0A2S2P7D1 A0A0J7KQY4 A0A0J7NJI3 A0A2S2NYY3 J9KLZ3 A0A0E3W2C5 A0A1W7R6C5 T1DG59 A0A0J7KIS5 T1DG89 A0A2M4BC18 Q9N9Z1 A0A2M4BC24 J9M6A5 K7JP45 Q868S0 V9GZD9 V5G2Q2 Q868S8 A0A2M4CJ33 A0A2M4CSA9 J9JZR8 J9M402 Q868S2 J9LQJ9 J9LB02 J9KT61 A0A2M4BD65 A0A0J7MYL9 A0A2M4AK35
A0A0J7N1D9 A0A0J7KS67 A0A0J7N7S8 A0A0J7KLT9 A0A0J7KIY5 A0A2S2N8I2 A0A2S2PBP5 A0A0J7KYD2 X1WJY4 A0A2S2PEZ7 A0A0J7JZ13 A0A0J7NEG9 A0A0J7NAV8 A0A2S2NJ82 A0A2H8TIE5 A0A0J7KL06 A0A0J7KMG5 J9JWJ3 A0A0J7MY17 A0A0J7NMN5 J9LVU7 A0A2S2P488 X1WT13 J9KNS1 A0A2H8TR36 X1WMW4 A0A2S2NQQ6 X1WXT5 J9KNG8 A0A2S2PEU2 J9KNN8 A0A2S2PRL8 T1DCM0 A0A142LX45 X1WSG8 A0A2S2P7D1 A0A0J7KQY4 A0A0J7NJI3 A0A2S2NYY3 J9KLZ3 A0A0E3W2C5 A0A1W7R6C5 T1DG59 A0A0J7KIS5 T1DG89 A0A2M4BC18 Q9N9Z1 A0A2M4BC24 J9M6A5 K7JP45 Q868S0 V9GZD9 V5G2Q2 Q868S8 A0A2M4CJ33 A0A2M4CSA9 J9JZR8 J9M402 Q868S2 J9LQJ9 J9LB02 J9KT61 A0A2M4BD65 A0A0J7MYL9 A0A2M4AK35
EMBL
AB090825
BAC57926.1
AB078935
BAC06462.1
AB078930
BAC06454.1
+ More
AB078931 BAC06456.1 AB078929 BAC06452.1 KZ149896 PZC78733.1 LBMM01012027 KMQ86470.1 LBMM01003679 KMQ93242.1 LBMM01008682 KMQ88705.1 LBMM01005733 KMQ91252.1 LBMM01006954 KMQ90166.1 GGMR01000779 MBY13398.1 GGMR01014195 MBY26814.1 LBMM01002052 KMQ95336.1 ABLF02011238 GGMR01015159 MBY27778.1 LBMM01019839 KMQ83438.1 LBMM01006110 KMQ90925.1 LBMM01007425 KMQ89740.1 GGMR01004600 MBY17219.1 GFXV01002090 MBW13895.1 LBMM01006063 KMQ90967.1 LBMM01005465 KMQ91492.1 ABLF02013358 ABLF02013361 ABLF02054869 LBMM01014122 KMQ85315.1 LBMM01003245 KMQ93755.1 ABLF02034925 GGMR01011682 MBY24301.1 ABLF02014378 ABLF02018808 GFXV01003903 MBW15708.1 ABLF02059310 GGMR01006838 MBY19457.1 ABLF02004854 ABLF02041317 ABLF02005203 ABLF02041316 GGMR01015354 MBY27973.1 ABLF02023257 GGMR01019493 MBY32112.1 GALA01001777 JAA93075.1 KU543683 AMS38371.1 ABLF02004597 ABLF02041324 GGMR01012748 MBY25367.1 LBMM01004262 KMQ92649.1 LBMM01004261 KMQ92650.1 GGMR01009778 MBY22397.1 ABLF02023401 ABLF02029423 ABLF02060278 HACL01000281 CFW94575.1 GEHC01000964 JAV46681.1 GALA01000302 JAA94550.1 LBMM01006977 KMQ90149.1 GALA01001776 JAA93076.1 GGFJ01001453 MBW50594.1 AJ278684 CAB99192.1 GGFJ01001454 MBW50595.1 ABLF02020969 AAZX01001144 AB090817 BAC57910.1 M93691 AAA29365.1 GALX01004171 JAB64295.1 AB090813 BAC57902.1 GGFL01001192 MBW65370.1 GGFL01004046 MBW68224.1 ABLF02009709 ABLF02009711 ABLF02028779 AB090816 BAC57908.1 ABLF02018471 ABLF02058352 ABLF02011136 ABLF02014426 ABLF02009073 ABLF02030702 ABLF02042963 GGFJ01001841 MBW50982.1 LBMM01013668 KMQ85535.1 GGFK01007771 MBW41092.1
AB078931 BAC06456.1 AB078929 BAC06452.1 KZ149896 PZC78733.1 LBMM01012027 KMQ86470.1 LBMM01003679 KMQ93242.1 LBMM01008682 KMQ88705.1 LBMM01005733 KMQ91252.1 LBMM01006954 KMQ90166.1 GGMR01000779 MBY13398.1 GGMR01014195 MBY26814.1 LBMM01002052 KMQ95336.1 ABLF02011238 GGMR01015159 MBY27778.1 LBMM01019839 KMQ83438.1 LBMM01006110 KMQ90925.1 LBMM01007425 KMQ89740.1 GGMR01004600 MBY17219.1 GFXV01002090 MBW13895.1 LBMM01006063 KMQ90967.1 LBMM01005465 KMQ91492.1 ABLF02013358 ABLF02013361 ABLF02054869 LBMM01014122 KMQ85315.1 LBMM01003245 KMQ93755.1 ABLF02034925 GGMR01011682 MBY24301.1 ABLF02014378 ABLF02018808 GFXV01003903 MBW15708.1 ABLF02059310 GGMR01006838 MBY19457.1 ABLF02004854 ABLF02041317 ABLF02005203 ABLF02041316 GGMR01015354 MBY27973.1 ABLF02023257 GGMR01019493 MBY32112.1 GALA01001777 JAA93075.1 KU543683 AMS38371.1 ABLF02004597 ABLF02041324 GGMR01012748 MBY25367.1 LBMM01004262 KMQ92649.1 LBMM01004261 KMQ92650.1 GGMR01009778 MBY22397.1 ABLF02023401 ABLF02029423 ABLF02060278 HACL01000281 CFW94575.1 GEHC01000964 JAV46681.1 GALA01000302 JAA94550.1 LBMM01006977 KMQ90149.1 GALA01001776 JAA93076.1 GGFJ01001453 MBW50594.1 AJ278684 CAB99192.1 GGFJ01001454 MBW50595.1 ABLF02020969 AAZX01001144 AB090817 BAC57910.1 M93691 AAA29365.1 GALX01004171 JAB64295.1 AB090813 BAC57902.1 GGFL01001192 MBW65370.1 GGFL01004046 MBW68224.1 ABLF02009709 ABLF02009711 ABLF02028779 AB090816 BAC57908.1 ABLF02018471 ABLF02058352 ABLF02011136 ABLF02014426 ABLF02009073 ABLF02030702 ABLF02042963 GGFJ01001841 MBW50982.1 LBMM01013668 KMQ85535.1 GGFK01007771 MBW41092.1
Proteomes
Pfam
Interpro
Gene 3D
ProteinModelPortal
Q868Q4
Q8MY27
Q8MY33
Q8MY31
Q8MY35
A0A2W1C3J5
+ More
A0A0J7N1D9 A0A0J7KS67 A0A0J7N7S8 A0A0J7KLT9 A0A0J7KIY5 A0A2S2N8I2 A0A2S2PBP5 A0A0J7KYD2 X1WJY4 A0A2S2PEZ7 A0A0J7JZ13 A0A0J7NEG9 A0A0J7NAV8 A0A2S2NJ82 A0A2H8TIE5 A0A0J7KL06 A0A0J7KMG5 J9JWJ3 A0A0J7MY17 A0A0J7NMN5 J9LVU7 A0A2S2P488 X1WT13 J9KNS1 A0A2H8TR36 X1WMW4 A0A2S2NQQ6 X1WXT5 J9KNG8 A0A2S2PEU2 J9KNN8 A0A2S2PRL8 T1DCM0 A0A142LX45 X1WSG8 A0A2S2P7D1 A0A0J7KQY4 A0A0J7NJI3 A0A2S2NYY3 J9KLZ3 A0A0E3W2C5 A0A1W7R6C5 T1DG59 A0A0J7KIS5 T1DG89 A0A2M4BC18 Q9N9Z1 A0A2M4BC24 J9M6A5 K7JP45 Q868S0 V9GZD9 V5G2Q2 Q868S8 A0A2M4CJ33 A0A2M4CSA9 J9JZR8 J9M402 Q868S2 J9LQJ9 J9LB02 J9KT61 A0A2M4BD65 A0A0J7MYL9 A0A2M4AK35
A0A0J7N1D9 A0A0J7KS67 A0A0J7N7S8 A0A0J7KLT9 A0A0J7KIY5 A0A2S2N8I2 A0A2S2PBP5 A0A0J7KYD2 X1WJY4 A0A2S2PEZ7 A0A0J7JZ13 A0A0J7NEG9 A0A0J7NAV8 A0A2S2NJ82 A0A2H8TIE5 A0A0J7KL06 A0A0J7KMG5 J9JWJ3 A0A0J7MY17 A0A0J7NMN5 J9LVU7 A0A2S2P488 X1WT13 J9KNS1 A0A2H8TR36 X1WMW4 A0A2S2NQQ6 X1WXT5 J9KNG8 A0A2S2PEU2 J9KNN8 A0A2S2PRL8 T1DCM0 A0A142LX45 X1WSG8 A0A2S2P7D1 A0A0J7KQY4 A0A0J7NJI3 A0A2S2NYY3 J9KLZ3 A0A0E3W2C5 A0A1W7R6C5 T1DG59 A0A0J7KIS5 T1DG89 A0A2M4BC18 Q9N9Z1 A0A2M4BC24 J9M6A5 K7JP45 Q868S0 V9GZD9 V5G2Q2 Q868S8 A0A2M4CJ33 A0A2M4CSA9 J9JZR8 J9M402 Q868S2 J9LQJ9 J9LB02 J9KT61 A0A2M4BD65 A0A0J7MYL9 A0A2M4AK35
PDB
6AR3
E-value=5.58581e-06,
Score=122
Ontologies
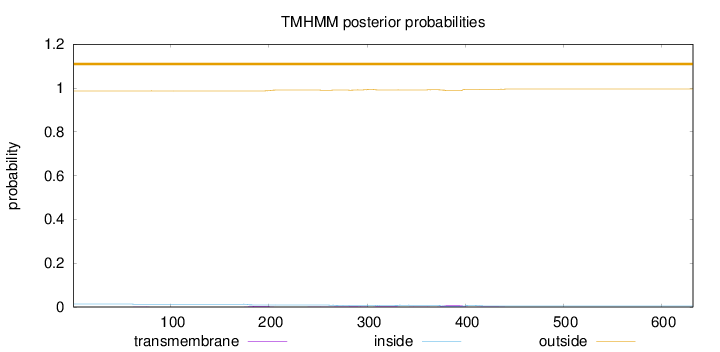
Topology
Length:
632
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.48976
Exp number, first 60 AAs:
0.00014
Total prob of N-in:
0.01326
outside
1 - 632
Population Genetic Test Statistics
Pi
176.122451
Theta
25.838699
Tajima's D
2.031468
CLR
148.530677
CSRT
0.889905504724764
Interpretation
Uncertain
